Note
Click here to download the full example code
Parameter space¶
In this example, we will see the basics of ParameterSpace.
from __future__ import absolute_import, division, print_function, unicode_literals
from future import standard_library
from gemseo.algos.parameter_space import ParameterSpace
from gemseo.api import configure_logger, create_discipline, create_scenario
configure_logger()
standard_library.install_aliases()
Firstly, a ParameterSpace does not require any mandatory argument.
Create a parameter space¶
parameter_space = ParameterSpace()
Then, we can add either deterministic variables from their lower and upper
bounds (use DesignSpace.add_variable())
or uncertain variables from their distribution names and parameters
(use add_random_variable())
parameter_space.add_variable("x", l_b=-2.0, u_b=2.0)
parameter_space.add_random_variable("y", "SPNormalDistribution", mu=0.0, sigma=1.0)
print(parameter_space)
Out:
+-------------------------------------------------------------------------------------------------------------------------------------------------------------------+
| Parameter space |
+------+-------------+-------+-------------+-------+-------------------------+----------------+-------------+------+--------------------+---------------------------+
| name | lower_bound | value | upper_bound | type | Initial distribution | Transformation | Support | Mean | Standard deviation | Range |
+------+-------------+-------+-------------+-------+-------------------------+----------------+-------------+------+--------------------+---------------------------+
| x | -2 | None | 2 | float | | | | | | |
| y | -inf | 0 | inf | float | norm(mu=0.0, sigma=1.0) | y | [-inf inf] | 0.0 | 1.0 | [-7.03448383 7.03448691] |
+------+-------------+-------+-------------+-------+-------------------------+----------------+-------------+------+--------------------+---------------------------+
We can check that the deterministic and uncertain variables are implemented as deterministic and deterministic variables respectively:
print("x is deterministic: ", parameter_space.is_deterministic("x"))
print("y is deterministic: ", parameter_space.is_deterministic("y"))
print("x is uncertain: ", parameter_space.is_uncertain("x"))
print("y is uncertain: ", parameter_space.is_uncertain("y"))
Out:
x is deterministic: True
y is deterministic: False
x is uncertain: False
y is uncertain: True
Sample from the parameter space¶
We can sample the uncertain variables from the ParameterSpace and
get values either as an array (default value) or as a dictionary:
sample = parameter_space.get_sample(n_samples=2, as_dict=True)
print(sample)
sample = parameter_space.get_sample(n_samples=4)
print(sample)
Out:
[{'y': array([1.62434536])}, {'y': array([-0.61175641])}]
[[-0.52817175]
[-1.07296862]
[ 0.86540763]
[-2.3015387 ]]
Sample a discipline over the parameter space¶
We can also sample a discipline over the parameter space. For simplicity,
we instantiate an AnalyticDiscipline from a dictionary of
expressions and update the cache policy # so as to cache all data in memory.
discipline = create_discipline("AnalyticDiscipline", expressions_dict={"z": "x+y"})
discipline.set_cache_policy(discipline.MEMORY_FULL_CACHE)
From these parameter space and discipline, we build a DOEScenario
and execute it with a Latin Hypercube Sampling algorithm and 100 samples.
Warning
A Scenario deals with all variables available in the
DesignSpace. By inheritance, a DOEScenario deals
with all variables available in the ParameterSpace.
Thus, if we do not filter the uncertain variables, the
DOEScenario will consider all variables. In particular, the
deterministic variables will be consider as uniformly distributed.
scenario = create_scenario(
[discipline], "DisciplinaryOpt", "z", parameter_space, scenario_type="DOE"
)
scenario.execute({"algo": "lhs", "n_samples": 100})
Out:
{'eval_jac': False, 'algo': 'lhs', 'n_samples': 100}
We can visualize the result by encapsulating the disciplinary cache in
a Dataset:
dataset = discipline.cache.export_to_dataset()
This visualization can be tabular for example:
print(dataset.export_to_dataframe())
Out:
inputs outputs
x y z
0 0 0
0 1.299635 -0.989742 0.309892
1 -1.735541 -0.775826 -2.511367
2 -1.970047 -1.937612 -3.907659
3 1.069838 0.119366 1.189204
4 0.941424 1.992783 2.934207
.. ... ... ...
95 -0.209952 -1.634437 -1.844389
96 0.192194 -1.225602 -1.033407
97 0.672494 0.361084 1.033578
98 1.132283 0.913791 2.046074
99 -0.764176 0.263038 -0.501139
[100 rows x 3 columns]
or graphical by means of a scatter plot matrix for example:
dataset.plot("ScatterMatrix")
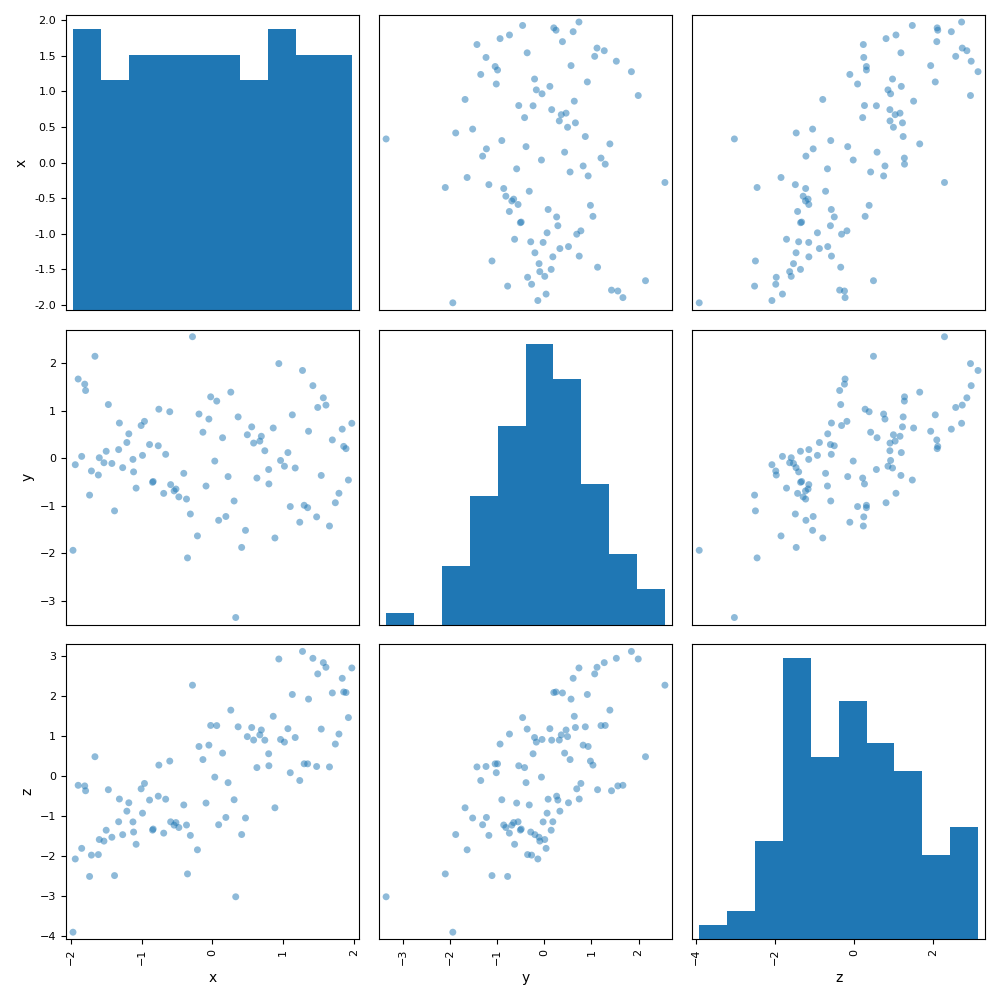
Out:
<gemseo.post.dataset.scatter_plot_matrix.ScatterMatrix object at 0x7fc29e072bb0>
Sample a discipline over the uncertain space¶
If we want to sample a discipline over the uncertain space, we need to filter the uncertain variables:
parameter_space.filter(parameter_space.uncertain_variables)
Out:
<gemseo.algos.parameter_space.ParameterSpace object at 0x7fc29b2718b0>
Then, we clear the cache, create a new scenario from this parameter space containing only the uncertain variables and execute it.
discipline.cache.clear()
scenario = create_scenario(
[discipline], "DisciplinaryOpt", "z", parameter_space, scenario_type="DOE"
)
scenario.execute({"algo": "lhs", "n_samples": 100})
Out:
{'eval_jac': False, 'algo': 'lhs', 'n_samples': 100}
Finally, we build a dataset from the disciplinary cache and visualize it. We can see that the deterministic variable ‘x’ is set to its default value for all evaluations, contrary to the previous case where we were considering the whole parameter space.
dataset = discipline.cache.export_to_dataset()
print(dataset.export_to_dataframe())
Out:
inputs outputs
x y z
0 0 0
0 0.0 0.415113 0.415113
1 0.0 0.055256 0.055256
2 0.0 0.489919 0.489919
3 0.0 -0.523228 -0.523228
4 0.0 1.138853 1.138853
.. ... ... ...
95 0.0 0.607114 0.607114
96 0.0 0.550599 0.550599
97 0.0 0.114359 0.114359
98 0.0 -0.382169 -0.382169
99 0.0 0.011416 0.011416
[100 rows x 3 columns]
Total running time of the script: ( 0 minutes 1.511 seconds)
