Note
Go to the end to download the full example code
Hybrid Jacobi/Newton MDA¶
from __future__ import annotations
from gemseo import configure_logger
from gemseo import create_discipline
from gemseo import create_mda
configure_logger()
<RootLogger root (INFO)>
Define a way to display results¶
def display_result(res, mda_name) -> None:
"""Display coupling and output variables in logger.
Args:
res: result of MDA
mda_name: name of the current MDA
"""
# names of the coupling variables
coupling_names = [
"y_11",
"y_12",
"y_14",
"y_21",
"y_23",
"y_24",
"y_31",
"y_32",
"y_34",
]
for coupling_var in coupling_names:
print(f"{mda_name}, coupling variable {coupling_var}: {res[coupling_var]}")
# names of the output variables
output_names = ["y_1", "y_2", "y_3", "y_4", "g_1", "g_2", "g_3"]
for output_name in output_names:
print(
f"{mda_name}, output variable {output_name}: {res[output_name]}",
)
Create, execute and post-process MDA¶
We do not need to specify the inputs, the default inputs of the MDA will be used and computed from the Default inputs of the disciplines
disciplines = create_discipline([
"SobieskiStructure",
"SobieskiPropulsion",
"SobieskiAerodynamics",
"SobieskiMission",
])
mda1 = create_mda("MDAJacobi", disciplines, max_mda_iter=1)
mda2 = create_mda("MDAGaussSeidel", disciplines)
mda_sequence = [mda1, mda2]
mda = create_mda("MDASequential", disciplines, mda_sequence=mda_sequence)
res = mda.execute()
display_result(res, mda.name)
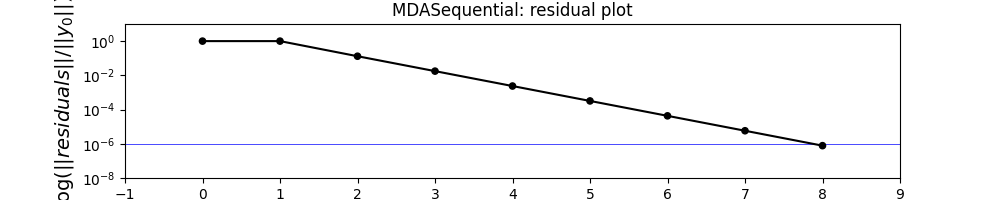
mda.plot_residual_history(
n_iterations=10,
logscale=[1e-8, 10.0],
save=False,
show=True,
fig_size=(10, 2),
)
WARNING - 09:00:07: MDAJacobi has reached its maximum number of iterations but the normed residual 1.0 is still above the tolerance 1e-06.
MDASequential, coupling variable y_11: [0.15591864]
MDASequential, coupling variable y_12: [5.06070636e+04 9.50000000e-01]
MDASequential, coupling variable y_14: [50607.06364014 7306.20262124]
MDASequential, coupling variable y_21: [50607.06364014]
MDASequential, coupling variable y_23: [12194.40917865]
MDASequential, coupling variable y_24: [4.15002178]
MDASequential, coupling variable y_31: [6354.40199361]
MDASequential, coupling variable y_32: [0.50280211]
MDASequential, coupling variable y_34: [1.10754577]
MDASequential, output variable y_1: [5.06070636e+04 7.30620262e+03 9.50000000e-01]
MDASequential, output variable y_2: [5.06070636e+04 1.21944092e+04 4.15002178e+00]
MDASequential, output variable y_3: [1.10754577e+00 6.35440199e+03 5.02802105e-01]
MDASequential, output variable y_4: [535.7821319]
MDASequential, output variable g_1: [ 0.035 -0.00666667 -0.0275 -0.04 -0.04833333 -0.09
-0.15 ]
MDASequential, output variable g_2: [-0.04]
MDASequential, output variable g_3: [-0.99719789 -0.00280211 0.16206032 -0.02 ]
INFO - 09:00:07: Requested 10 iterations but the residual history contains only 9, plotting all the residual history.
<Figure size 1000x200 with 1 Axes>
Total running time of the script: (0 minutes 0.265 seconds)
