Self-Organizing Maps¶
Preliminaries: instantiation and execution of the MDO scenario¶
Let’s start with the following code lines which instantiate and execute the MDOScenario :
from gemseo.api import create_discipline, create_scenario
formulation = 'MDF'
disciplines = create_discipline(["SobieskiPropulsion", "SobieskiAerodynamics",
"SobieskiMission", "SobieskiStructure"])
scenario = create_scenario(disciplines,
formulation=formulation,
objective_name="y_4",
maximize_objective=True,
design_space="design_space.txt")
scenario.set_differentiation_method("user")
algo_options = {'max_iter': 10, 'algo': "NLOPT_COBYLA"}
for constraint in ["g_1","g_2","g_3"]:
scenario.add_constraint(constraint, 'ineq')
scenario.execute(algo_options)
SOM¶
Description¶
The SOM post processing perform a Self Organizing Map clustering
on optimization history.
A SOM is a 2D representation of a design of experiments
which requires dimensionality reduction since it may be in very high dimension.
Options of the plot method are the figure width and height,
and the x- and y- number of cells in the SOM.
It is also possible either to save the plot, to show the plot or both.
Options¶
annotate,
Unknown- add label of neuron value to SOM plotextension,
str- file extensionfile_path,
str- the base paths of the files to exportheight,
Unknown- figure heightn_x,
int- x-sizen_y,
int- y-sizesave,
bool- if True, exports plot to pdfshow,
bool- if True, displays the plot windowswidth,
Unknown- figure width
Case of the MDF formulation¶
To plot the Self-Organizing Maps, use the API method execute_post()
with the keyword “SOM”, the new dimension n_x and n_y and
additional arguments concerning the type of display (file, screen, both):
scenario.post_process(“SOM”, save=False, n_x=4, n_y=4, show=True)
A SOM is built by using an unsupervised artificial neural network [KSH01].
A map of size n_x.n_y is generated, where
n_x is the number of neurons in the \(x\) direction and n_y
is the number of neurons in the \(y\) direction. The design space
(whatever the dimension) is reduced to a 2D representation based on
n_x.n_y neurons. Samples are clustered to a neuron when their design
variables are close in terms of L2 norm. A neuron is always located at the same place on a
map. Each neuron is colored according to the average value for a given
criterion. This helps to qualitatively analyze if parts of the design
space are good according to some criteria and not for others, and where
compromises should be made. A white neuron has no sample associated with
it: not enough evaluations were provided to train the SOM.
SOM provide a qualitative view of the objective function and the constraints, and of their relative behaviors.
Figure SOM example on the Sobieski problem illustrates a SOM on the Sobieski use case. The optimization method is a
(costly) derivative free algorithm (NLOPT_COBYLA), since relevant
are obtained at the cost of numerous evaluations of the functions. For
more details, please read the paper by
[KJO+06] on wing MDO post-processing
using SOM.
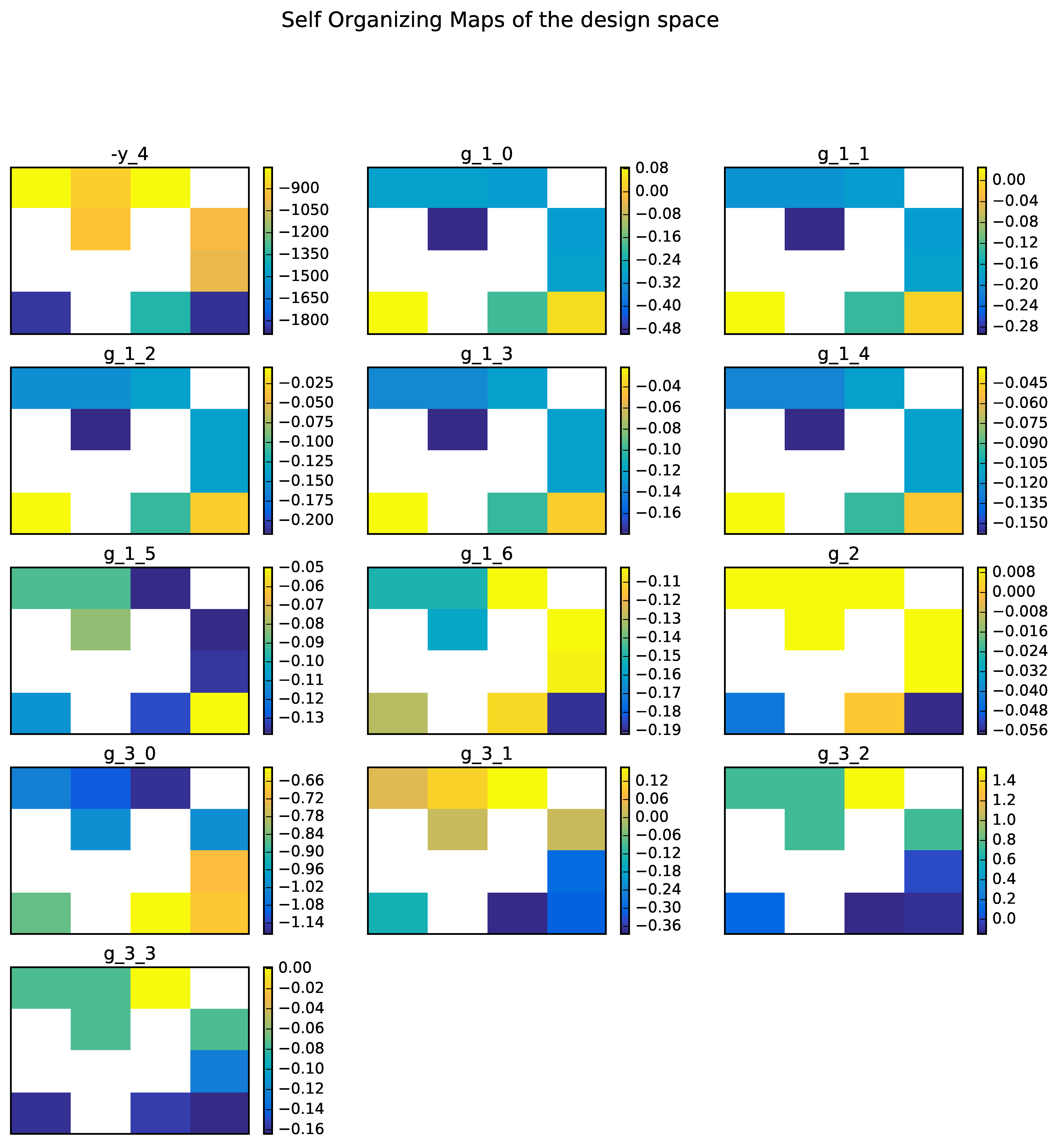
SOM example on the Sobieski problem¶
A DOE may also be a good way to produce SOM maps. In figure SOM example on the Sobieski problem with a 10 000 samples DOE is an example with 10000 points on the same test case. This produces more relevant SOM plots.
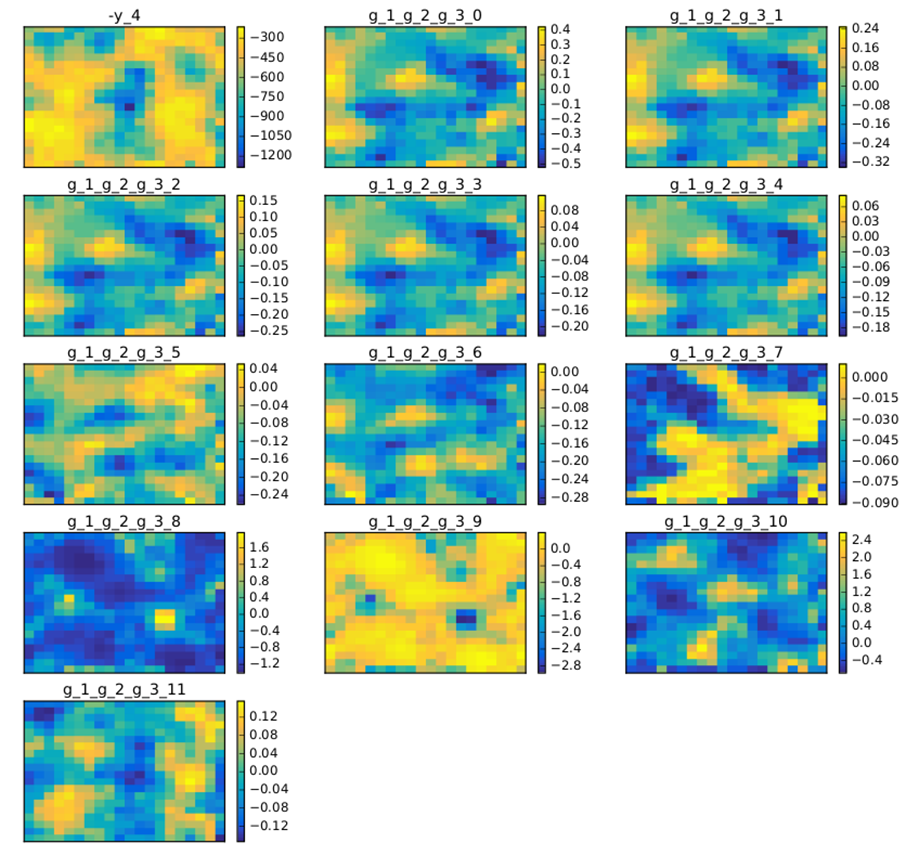
SOM example on the Sobieski problem with a 10 000 samples DOE¶
