Note
Click here to download the full example code
Quality measure for surrogate model comparison¶
In this example we use the quality measure class to compare the performances of a mixture of experts (MoE) and a random forest algorithm under different circumstances. We will consider two different datasets: A 1D function, and the Rosenbrock dataset (two inputs and one output).
Import¶
from __future__ import annotations
import matplotlib.pyplot as plt
from gemseo.api import configure_logger
from gemseo.api import load_dataset
from gemseo.core.dataset import Dataset
from gemseo.mlearning.api import create_regression_model
from gemseo.mlearning.qual_measure.mse_measure import MSEMeasure
from gemseo.mlearning.transform.scaler.min_max_scaler import MinMaxScaler
from numpy import hstack
from numpy import linspace
from numpy import meshgrid
from numpy import sin
configure_logger()
<RootLogger root (INFO)>
Test on 1D dataset¶
In this section we create a dataset from an analytical expression of a 1D function, and compare the errors of the two regression models.
Create 1D dataset from expression¶
def data_gen(x):
return 3 + 0.5 * sin(14 * x) * (x <= 0.7) + (x > 0.7) * (0.8 + 6 * (x - 1) ** 2)
x = linspace(0, 1, 25)
y = data_gen(x)
data = hstack((x[:, None], y[:, None]))
variables = ["x", "y"]
sizes = {"x": 1, "y": 1}
groups = {"x": Dataset.INPUT_GROUP, "y": Dataset.OUTPUT_GROUP}
dataset = Dataset("dataset_name")
dataset.set_from_array(data, variables, sizes, groups)
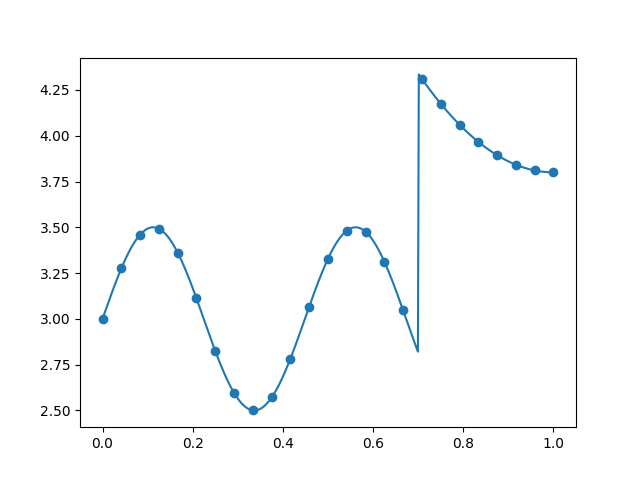
Plot 1D data¶
x_refined = linspace(0, 1, 500)
y_refined = data_gen(x_refined)
plt.plot(x_refined, y_refined)
plt.scatter(x, y)
plt.show()
Create regression algorithms¶
moe = create_regression_model(
"MOERegressor", dataset, transformer={"outputs": MinMaxScaler()}
)
moe.set_clusterer("GaussianMixture", n_components=4)
moe.set_classifier("KNNClassifier", n_neighbors=3)
moe.set_regressor(
"PolynomialRegressor", degree=5, l2_penalty_ratio=1, penalty_level=0.00005
)
randfor = create_regression_model(
"RandomForestRegressor",
dataset,
transformer={"outputs": MinMaxScaler()},
n_estimators=50,
)
Compute measures (Mean Squared Error)¶
measure_moe = MSEMeasure(moe)
measure_randfor = MSEMeasure(randfor)
Evaluate on training set directly (keyword: ‘learn’)¶
print("Learn:")
print("Error MoE:", measure_moe.evaluate(method="learn"))
print("Error Random Forest:", measure_randfor.evaluate(method="learn"))
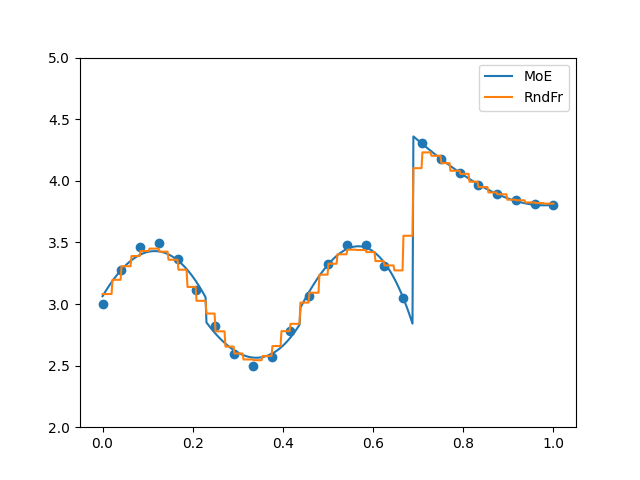
plt.figure()
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(x_refined, randfor.predict(x_refined[:, None]).flatten(), label="RndFr")
plt.scatter(x, y)
plt.legend()
plt.ylim(2, 5)
plt.show()
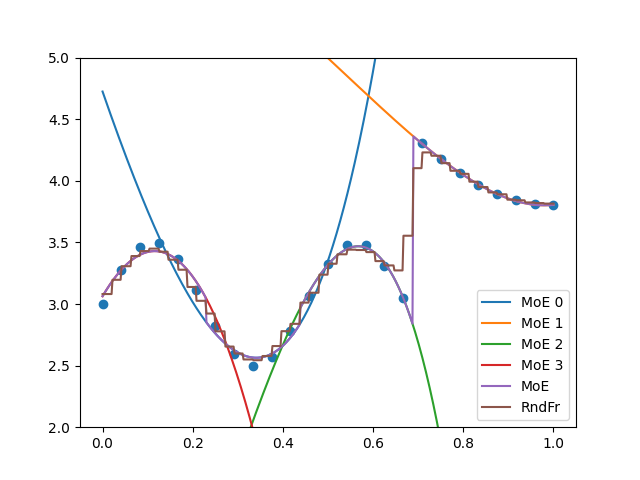
plt.figure()
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 0).flatten(), label="MoE 0"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 1).flatten(), label="MoE 1"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 2).flatten(), label="MoE 2"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 3).flatten(), label="MoE 3"
)
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(x_refined, randfor.predict(x_refined[:, None]).flatten(), label="RndFr")
plt.scatter(x, y)
plt.legend()
plt.ylim(2, 5)
plt.show()
Learn:
Error MoE: [0.00130025]
Error Random Forest: [0.01388602]
Evaluate using cross validation (keyword: ‘kfolds’)¶
In order to better consider the generalization error, perform a k-folds cross validation algorithm. We also plot the predictions from the last iteration of the algorithm.
print("K-folds:")
print("Error MoE:", measure_moe.evaluate("kfolds"))
print("Error Random Forest:", measure_randfor.evaluate("kfolds"))
print("Loo:")
print("Error MoE:", measure_moe.evaluate("loo"))
print("Error Random Forest:", measure_randfor.evaluate("loo"))
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(
x_refined, randfor.predict(x_refined[:, None]).flatten(), label="Random Forest"
)
plt.scatter(x, y)
plt.legend()
plt.show()
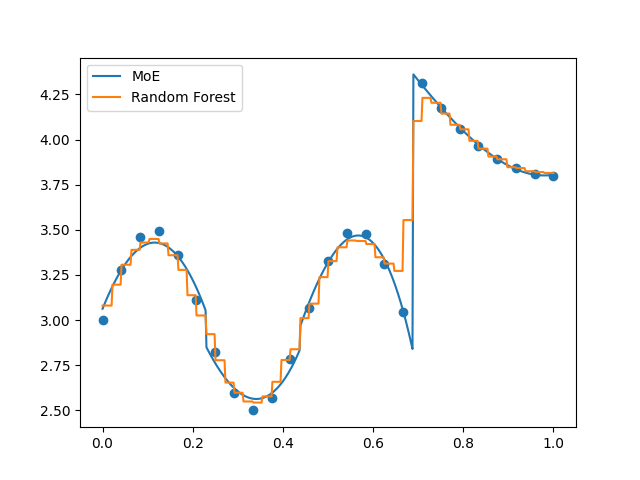
K-folds:
Error MoE: [0.00157915]
Error Random Forest: [0.00931296]
Loo:
Error MoE: [0.00221518]
Error Random Forest: [0.01085501]
Test on 2D dataset (Rosenbrock)¶
In this section, we load the Rosenbrock dataset, and compare the error measures for the two regression models.
Load dataset¶
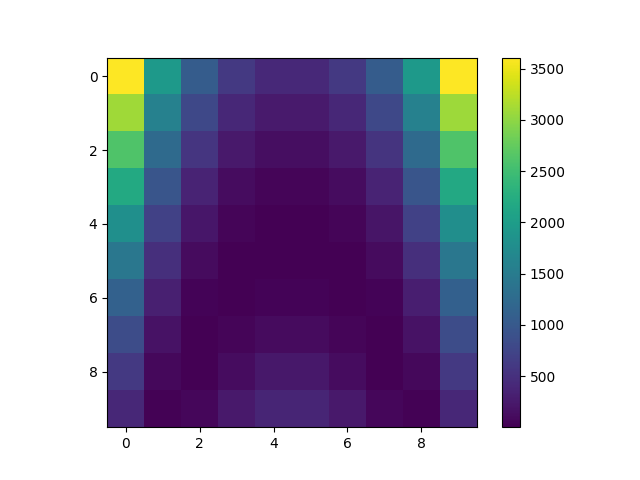
dataset = load_dataset("RosenbrockDataset", opt_naming=False)
x = dataset.get_data_by_group(dataset.INPUT_GROUP)
y = dataset.get_data_by_group(dataset.OUTPUT_GROUP)
Y = y.reshape((10, 10))
refinement = 100
x_refined = linspace(-2, 2, refinement)
X_1_refined, X_2_refined = meshgrid(x_refined, x_refined)
x_1_refined, x_2_refined = X_1_refined.flatten(), X_2_refined.flatten()
x_refined = hstack((x_1_refined[:, None], x_2_refined[:, None]))
print(dataset)
Rosenbrock
Number of samples: 100
Number of variables: 2
Variables names and sizes by group:
inputs: x (2)
outputs: rosen (1)
Number of dimensions (total = 3) by group:
inputs: 2
outputs: 1
Create regression algorithms¶
moe = create_regression_model(
"MOERegressor", dataset, transformer={"outputs": MinMaxScaler()}
)
moe.set_clusterer("KMeans", n_clusters=3)
moe.set_classifier("KNNClassifier", n_neighbors=5)
moe.set_regressor(
"PolynomialRegressor", degree=5, l2_penalty_ratio=1, penalty_level=0.1
)
randfor = create_regression_model(
"RandomForestRegressor",
dataset,
transformer={"outputs": MinMaxScaler()},
n_estimators=200,
)
Compute measures (Mean Squared Error)¶
measure_moe = MSEMeasure(moe)
measure_randfor = MSEMeasure(randfor)
print("Learn:")
print("Error MoE:", measure_moe.evaluate(method="learn"))
print("Error Random Forest:", measure_randfor.evaluate(method="learn"))
print("K-folds:")
print("Error MoE:", measure_moe.evaluate("kfolds"))
print("Error Random Forest:", measure_randfor.evaluate("kfolds"))
Learn:
Error MoE: [8.19866424]
Error Random Forest: [4567.22027112]
K-folds:
Error MoE: [26.99008863]
Error Random Forest: [17161.54269564]
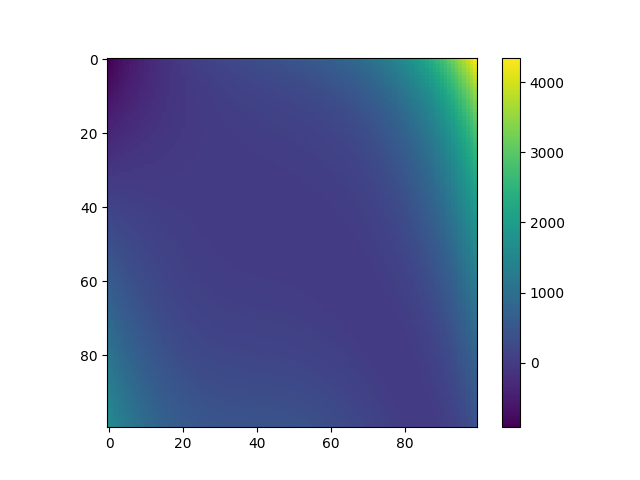
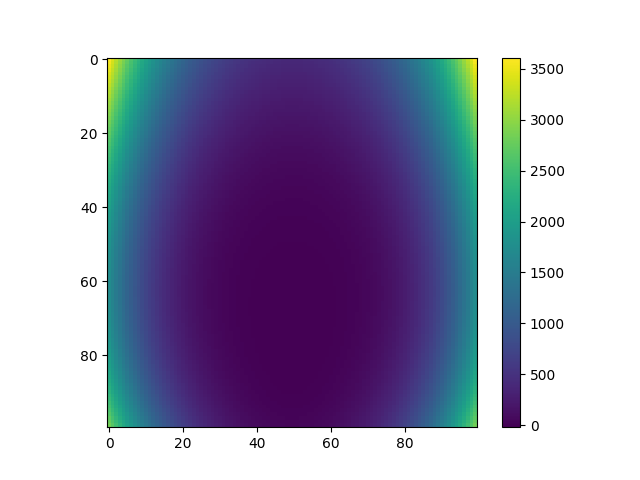
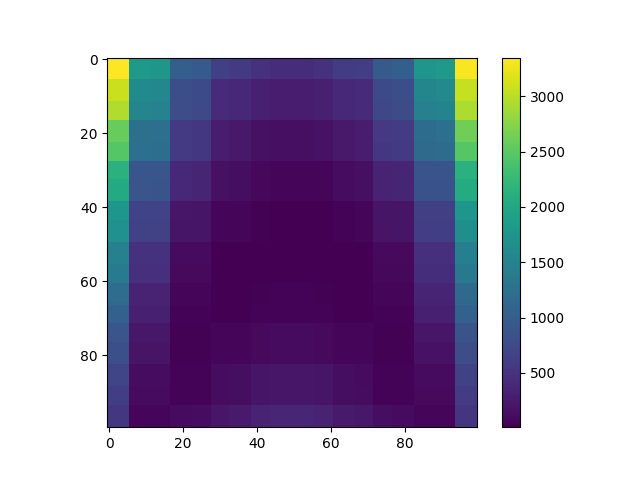
Plot data¶
plt.imshow(Y, interpolation="nearest")
plt.colorbar()
plt.show()
Plot predictions¶
moe.learn()
randfor.learn()
Y_pred_moe = moe.predict(x_refined).reshape((refinement, refinement))
Y_pred_moe_0 = moe.predict_local_model(x_refined, 0).reshape((refinement, refinement))
Y_pred_moe_1 = moe.predict_local_model(x_refined, 1).reshape((refinement, refinement))
Y_pred_moe_2 = moe.predict_local_model(x_refined, 2).reshape((refinement, refinement))
Y_pred_randfor = randfor.predict(x_refined).reshape((refinement, refinement))
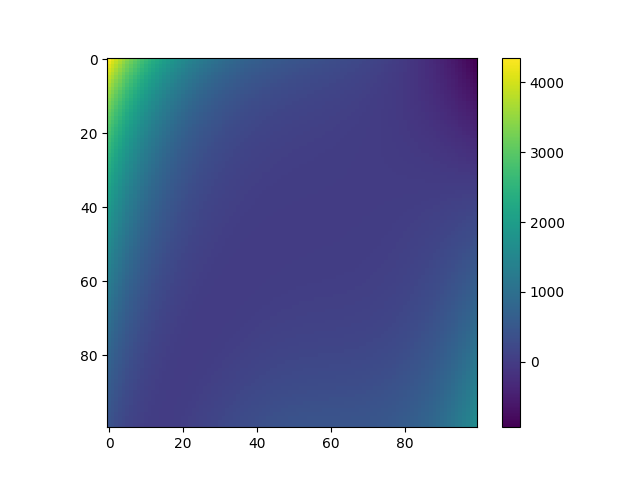
Plot mixture of experts predictions¶
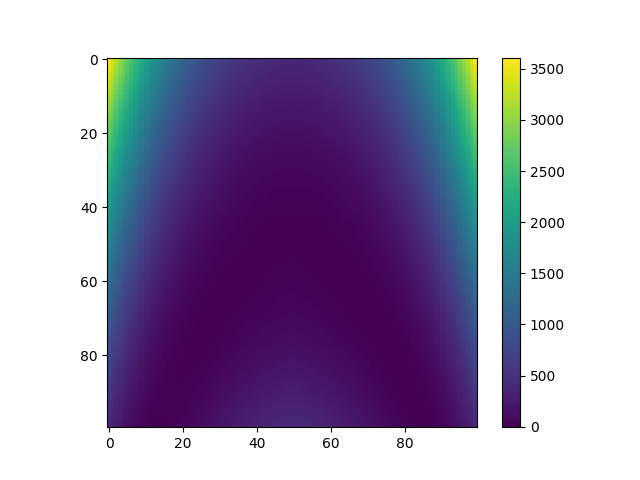
plt.imshow(Y_pred_moe)
plt.colorbar()
plt.show()
Plot local models¶
plt.figure()
plt.imshow(Y_pred_moe_0)
plt.colorbar()
plt.show()
plt.figure()
plt.imshow(Y_pred_moe_1)
plt.colorbar()
plt.show()
plt.figure()
plt.imshow(Y_pred_moe_2)
plt.colorbar()
plt.show()
Plot random forest predictions¶
plt.imshow(Y_pred_randfor)
plt.colorbar()
plt.show()
Total running time of the script: ( 0 minutes 5.988 seconds)