Note
Go to the end to download the full example code
Quality measure for surrogate model comparison¶
In this example we use the quality measure class to compare the performances of a mixture of experts (MoE) and a random forest algorithm under different circumstances. We will consider two different datasets: A 1D function, and the Rosenbrock dataset (two inputs and one output).
Import¶
from __future__ import annotations
import matplotlib.pyplot as plt
from gemseo import configure_logger
from gemseo import create_benchmark_dataset
from gemseo.datasets.io_dataset import IODataset
from gemseo.mlearning import create_regression_model
from gemseo.mlearning.quality_measures.mse_measure import MSEMeasure
from gemseo.mlearning.transformers.scaler.min_max_scaler import MinMaxScaler
from numpy import hstack
from numpy import linspace
from numpy import meshgrid
from numpy import sin
configure_logger()
<RootLogger root (INFO)>
Test on 1D dataset¶
In this section we create a dataset from an analytical expression of a 1D function, and compare the errors of the two regression models.
Create 1D dataset from expression¶
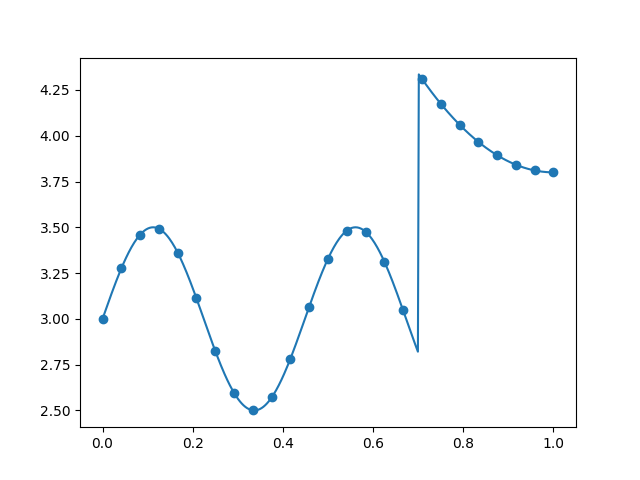
def data_gen(x):
return 3 + 0.5 * sin(14 * x) * (x <= 0.7) + (x > 0.7) * (0.8 + 6 * (x - 1) ** 2)
x = linspace(0, 1, 25)
y = data_gen(x)
data = hstack((x[:, None], y[:, None]))
variables = ["x", "y"]
sizes = {"x": 1, "y": 1}
groups = {"x": IODataset.INPUT_GROUP, "y": IODataset.OUTPUT_GROUP}
dataset = IODataset.from_array(data, variables, sizes, groups)
Plot 1D data¶
x_refined = linspace(0, 1, 500)
y_refined = data_gen(x_refined)
plt.plot(x_refined, y_refined)
plt.scatter(x, y)
plt.show()
Create regression algorithms¶
moe = create_regression_model(
"MOERegressor", dataset, transformer={"outputs": MinMaxScaler()}
)
moe.set_clusterer("GaussianMixture", n_components=4)
moe.set_classifier("KNNClassifier", n_neighbors=3)
moe.set_regressor(
"PolynomialRegressor", degree=5, l2_penalty_ratio=1, penalty_level=0.00005
)
randfor = create_regression_model(
"RandomForestRegressor",
dataset,
transformer={"outputs": MinMaxScaler()},
n_estimators=50,
)
Compute measures (Mean Squared Error)¶
measure_moe = MSEMeasure(moe)
measure_randfor = MSEMeasure(randfor)
Evaluate on training set directly (keyword: ‘learn’)¶
print("Learn:")
print("Error MoE:", measure_moe.evaluate_learn())
print("Error Random Forest:", measure_randfor.evaluate_learn())
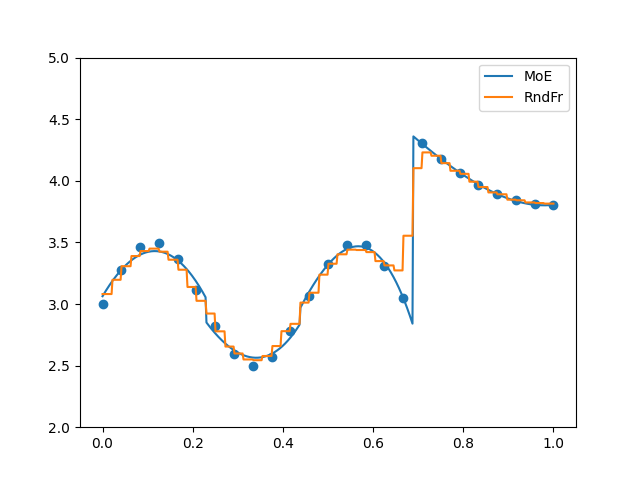
plt.figure()
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(x_refined, randfor.predict(x_refined[:, None]).flatten(), label="RndFr")
plt.scatter(x, y)
plt.legend()
plt.ylim(2, 5)
plt.show()
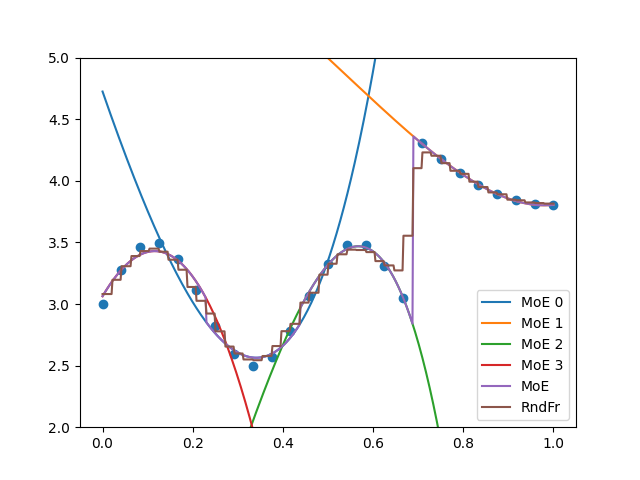
plt.figure()
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 0).flatten(), label="MoE 0"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 1).flatten(), label="MoE 1"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 2).flatten(), label="MoE 2"
)
plt.plot(
x_refined, moe.predict_local_model(x_refined[:, None], 3).flatten(), label="MoE 3"
)
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(x_refined, randfor.predict(x_refined[:, None]).flatten(), label="RndFr")
plt.scatter(x, y)
plt.legend()
plt.ylim(2, 5)
plt.show()
Learn:
Error MoE: [0.00130025]
Error Random Forest: [0.01388602]
Evaluate using cross validation (keyword: ‘kfolds’)¶
In order to better consider the generalization error, perform a k-folds cross validation algorithm. We also plot the predictions from the last iteration of the algorithm.
print("K-folds:")
print("Error MoE:", measure_moe.evaluate_kfolds())
print("Error Random Forest:", measure_randfor.evaluate_kfolds())
print("Loo:")
print("Error MoE:", measure_moe.evaluate_loo())
print("Error Random Forest:", measure_randfor.evaluate_loo())
plt.plot(x_refined, moe.predict(x_refined[:, None]).flatten(), label="MoE")
plt.plot(
x_refined, randfor.predict(x_refined[:, None]).flatten(), label="Random Forest"
)
plt.scatter(x, y)
plt.legend()
plt.show()
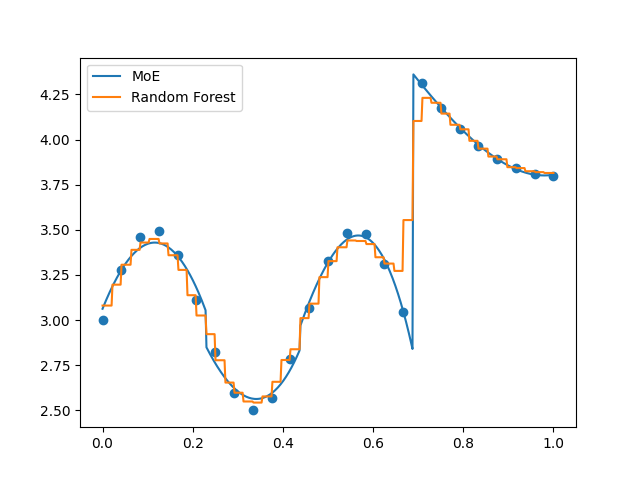
K-folds:
Error MoE: [0.00157915]
Error Random Forest: [0.00931296]
Loo:
Error MoE: [0.00221518]
Error Random Forest: [0.01085501]
Test on 2D dataset (Rosenbrock)¶
In this section, we load the Rosenbrock dataset, and compare the error measures for the two regression models.
Load dataset¶
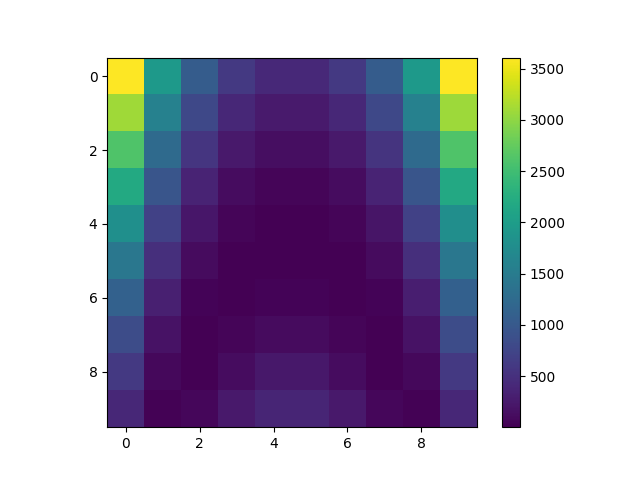
dataset = create_benchmark_dataset("RosenbrockDataset", opt_naming=False)
x = dataset.input_dataset.to_numpy()
y = dataset.output_dataset.to_numpy()
Y = y.reshape((10, 10))
refinement = 100
x_refined = linspace(-2, 2, refinement)
X_1_refined, X_2_refined = meshgrid(x_refined, x_refined)
x_1_refined, x_2_refined = X_1_refined.flatten(), X_2_refined.flatten()
x_refined = hstack((x_1_refined[:, None], x_2_refined[:, None]))
print(dataset)
GROUP inputs outputs
VARIABLE x rosen
COMPONENT 0 1 0
0 -2.000000 -2.0 3609.000000
1 -1.555556 -2.0 1959.952599
2 -1.111111 -2.0 1050.699741
3 -0.666667 -2.0 600.308642
4 -0.222222 -2.0 421.490779
.. ... ... ...
95 0.222222 2.0 381.095717
96 0.666667 2.0 242.086420
97 1.111111 2.0 58.600975
98 1.555556 2.0 17.927907
99 2.000000 2.0 401.000000
[100 rows x 3 columns]
Create regression algorithms¶
moe = create_regression_model(
"MOERegressor", dataset, transformer={"outputs": MinMaxScaler()}
)
moe.set_clusterer("KMeans", n_clusters=3)
moe.set_classifier("KNNClassifier", n_neighbors=5)
moe.set_regressor(
"PolynomialRegressor", degree=5, l2_penalty_ratio=1, penalty_level=0.1
)
randfor = create_regression_model(
"RandomForestRegressor",
dataset,
transformer={"outputs": MinMaxScaler()},
n_estimators=200,
)
Compute measures (Mean Squared Error)¶
measure_moe = MSEMeasure(moe)
measure_randfor = MSEMeasure(randfor)
print("Learn:")
print("Error MoE:", measure_moe.evaluate_learn())
print("Error Random Forest:", measure_randfor.evaluate_learn())
print("K-folds:")
print("Error MoE:", measure_moe.evaluate_kfolds())
print("Error Random Forest:", measure_randfor.evaluate_kfolds())
Learn:
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
Error MoE: [8.19866424]
Error Random Forest: [4567.22027112]
K-folds:
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
Error MoE: [26.99008863]
Error Random Forest: [17161.54269564]
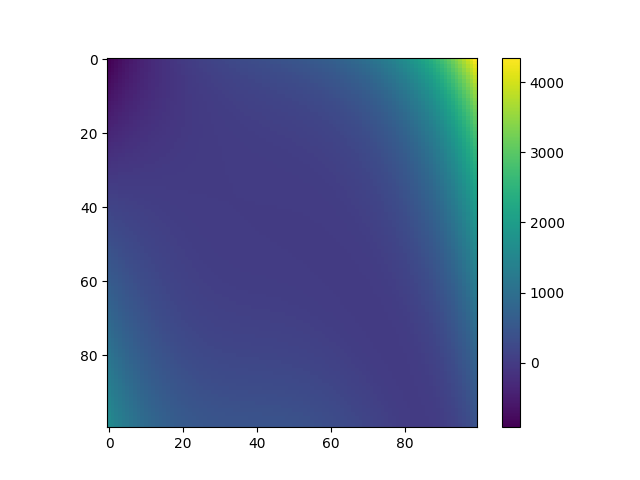
Plot data¶
plt.imshow(Y, interpolation="nearest")
plt.colorbar()
plt.show()
Plot predictions¶
moe.learn()
randfor.learn()
Y_pred_moe = moe.predict(x_refined).reshape((refinement, refinement))
Y_pred_moe_0 = moe.predict_local_model(x_refined, 0).reshape((refinement, refinement))
Y_pred_moe_1 = moe.predict_local_model(x_refined, 1).reshape((refinement, refinement))
Y_pred_moe_2 = moe.predict_local_model(x_refined, 2).reshape((refinement, refinement))
Y_pred_randfor = randfor.predict(x_refined).reshape((refinement, refinement))
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/5.0.0/lib/python3.9/site-packages/sklearn/cluster/_kmeans.py:870: FutureWarning: The default value of `n_init` will change from 10 to 'auto' in 1.4. Set the value of `n_init` explicitly to suppress the warning
warnings.warn(
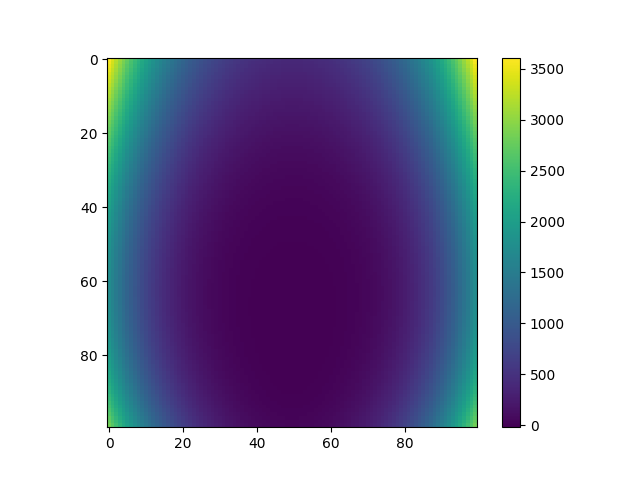
Plot mixture of experts predictions¶
plt.imshow(Y_pred_moe)
plt.colorbar()
plt.show()

Plot local models¶
plt.figure()
plt.imshow(Y_pred_moe_0)
plt.colorbar()
plt.show()
plt.figure()
plt.imshow(Y_pred_moe_1)
plt.colorbar()
plt.show()
plt.figure()
plt.imshow(Y_pred_moe_2)
plt.colorbar()
plt.show()
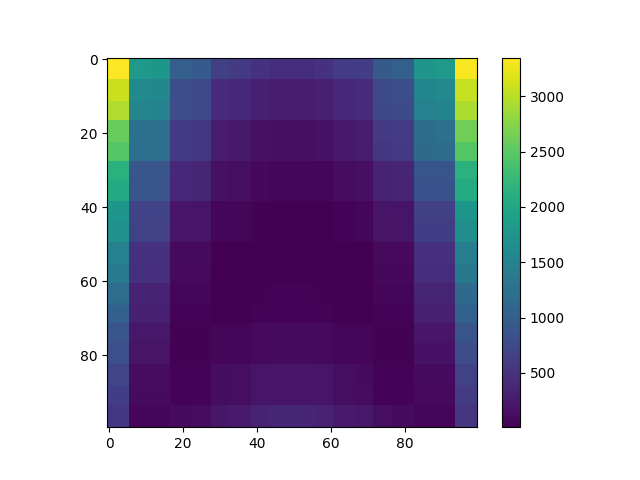
Plot random forest predictions¶
plt.imshow(Y_pred_randfor)
plt.colorbar()
plt.show()
Total running time of the script: ( 0 minutes 10.256 seconds)