Note
Go to the end to download the full example code
Parametric estimation of statistics¶
In this example, we want to estimate statistics from synthetic data. These data are 500 realizations of x_0, x_1, x_2 and x_3 distributed in the following way:
x_0: standard uniform distribution,
x_1: standard normal distribution,
x_2: standard Weibull distribution,
x_3: standard exponential distribution.
These samples are generated from the NumPy library.
from __future__ import annotations
from numpy import vstack
from numpy.random import default_rng
from gemseo import configure_logger
from gemseo import create_dataset
from gemseo.uncertainty import create_statistics
configure_logger()
<RootLogger root (INFO)>
Create synthetic data¶
rng = default_rng(0)
n_samples = 500
uniform_rand = rng.uniform(size=n_samples)
normal_rand = rng.normal(size=n_samples)
weibull_rand = rng.weibull(1.5, size=n_samples)
exponential_rand = rng.exponential(size=n_samples)
data = vstack((uniform_rand, normal_rand, weibull_rand, exponential_rand)).T
variables = ["x_0", "x_1", "x_2", "x_3"]
data
array([[ 6.36961687e-01, 1.35543803e+00, 1.24477385e-01,
1.91363961e-01],
[ 2.69786714e-01, 2.21160257e-03, 8.14109465e-01,
2.33137384e+00],
[ 4.09735239e-02, -7.90544810e-01, 4.64297251e-01,
5.53852517e-01],
...,
[ 9.85769635e-01, -1.16187331e+00, 9.62671893e-01,
8.66423178e-01],
[ 4.28024519e-01, 2.72032137e-01, 5.03249648e-01,
2.17492296e-01],
[ 8.43014715e-01, -7.66939588e-01, 8.18740909e-01,
8.75593057e-01]])
Create a ParametricStatistics object¶
We create a ParametricStatistics object
from this data encapsulated in a Dataset:
dataset = create_dataset("Dataset", data, variables)
and a list of names of candidate probability distributions:
exponential, normal and uniform distributions
(see ParametricStatistics.DistributionName).
We do not use the default
fitting criterion (‘BIC’) but ‘Kolmogorov’
(see ParametricStatistics.FittingCriterion
and ParametricStatistics.SignificanceTest).
tested_distributions = ["Exponential", "Normal", "Uniform"]
analysis = create_statistics(
dataset, tested_distributions=tested_distributions, fitting_criterion="Kolmogorov"
)
analysis
INFO - 13:52:41: Create ParametricStatistics(Dataset), a ParametricStatistics library.
INFO - 13:52:41: | Set goodness-of-fit criterion: Kolmogorov.
INFO - 13:52:41: | Set significance level of hypothesis test: 0.05.
INFO - 13:52:41: Fit different distributions (Exponential, Normal, Uniform) per variable and compute the goodness-of-fit criterion.
INFO - 13:52:41: | Fit different distributions for x_0.
INFO - 13:52:41: | Fit different distributions for x_1.
INFO - 13:52:41: | Fit different distributions for x_2.
INFO - 13:52:41: | Fit different distributions for x_3.
INFO - 13:52:41: Select the best distribution for each variable.
WARNING - 13:52:41: All criteria values are lower than the significance level 0.05.
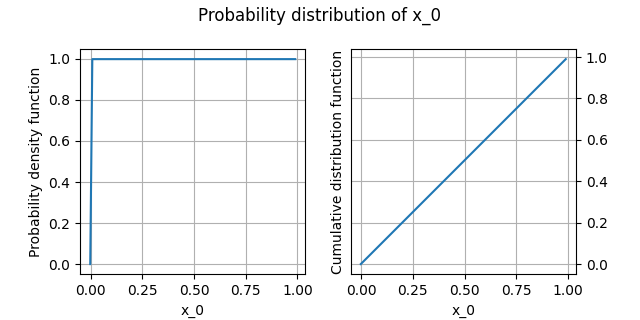
INFO - 13:52:41: | The best distribution for x_0[0] is Uniform([-0.00168518,0.999196]).
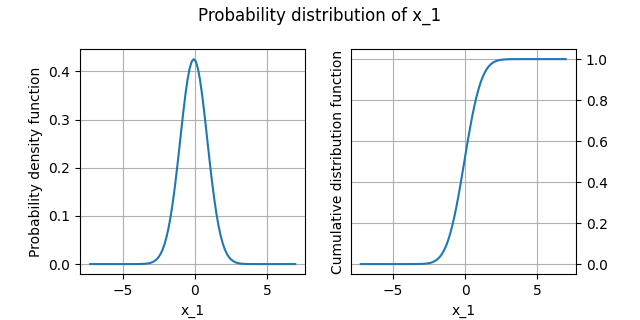
INFO - 13:52:41: | The best distribution for x_1[0] is Normal([-0.068321,0.937168]).
WARNING - 13:52:41: All criteria values are lower than the significance level 0.05.
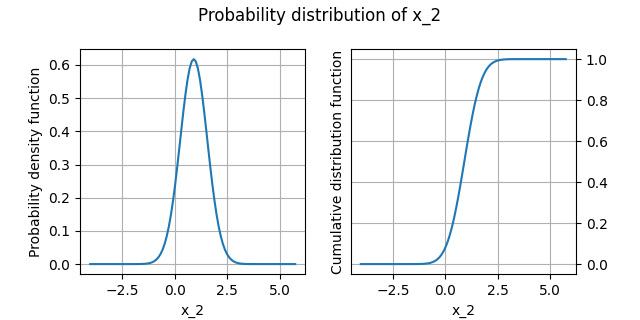
INFO - 13:52:41: | The best distribution for x_2[0] is Normal([0.905464,0.645926]).
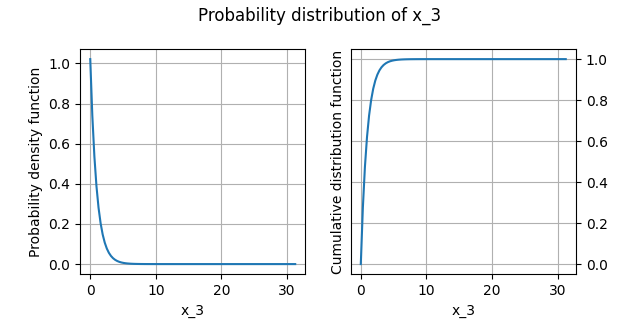
INFO - 13:52:41: | The best distribution for x_3[0] is Exponential([1.02161,0.00146538]).
Print the fitting matrix¶
At this step, an optimal distribution has been selected for each variable based on the tested distributions and on the Kolmogorov fitting criterion. We can print the fitting matrix to see the goodness-of-fit measures for each pair < variable, distribution > as well as the selected distribution for each variable. Note that in the case of significance tests, the goodness-of-fit measures are the p-values.
analysis.get_fitting_matrix()
'+----------+------------------------+------------------------+-------------------------+-------------+\n| Variable | Exponential | Normal | Uniform | Selection |\n+----------+------------------------+------------------------+-------------------------+-------------+\n| x_0 | 1.3319194027726772e-16 | 0.02244629355147776 | 0.035215303972709444 | Uniform |\n| x_1 | 1.177360930542324e-55 | 0.9894613754182425 | 2.773038784889384e-21 | Normal |\n| x_2 | 1.624812325803554e-08 | 0.00024649291266878377 | 8.697307474207096e-95 | Normal |\n| x_3 | 0.6841041353514807 | 1.1084730889960638e-13 | 1.0968178545541834e-160 | Exponential |\n+----------+------------------------+------------------------+-------------------------+-------------+'
One can also plot the tested distributions over an histogram of the data as well as the corresponding values of the Kolmogorov fitting criterion:
analysis.plot_criteria("x_0")
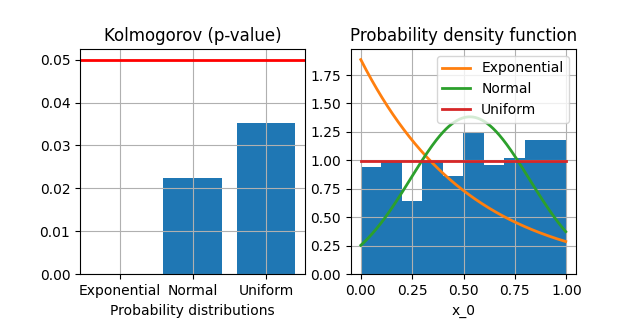
<Figure size 640x320 with 2 Axes>
Get statistics¶
From this ParametricStatistics instance,
we can easily get statistics for the different variables
based on the selected distributions.
Get minimum¶
Here is the minimum value for the different variables:
analysis.compute_minimum()
{'x_0': array([-0.00168518]), 'x_1': array([-inf]), 'x_2': array([-inf]), 'x_3': array([0.00146538])}
Get maximum¶
Here is the minimum value for the different variables:
analysis.compute_maximum()
{'x_0': array([0.99919581]), 'x_1': array([inf]), 'x_2': array([inf]), 'x_3': array([inf])}
Get range¶
Here is the range, i.e. the difference between the minimum and maximum values, for the different variables:
analysis.compute_range()
{'x_0': array([1.000881]), 'x_1': array([inf]), 'x_2': array([inf]), 'x_3': array([inf])}
Get mean¶
Here is the mean value for the different variables:
analysis.compute_mean()
{'x_0': array([0.49875531]), 'x_1': array([-0.06832097]), 'x_2': array([0.90546442]), 'x_3': array([0.98031191])}
Get standard deviation¶
Here is the standard deviation for the different variables:
analysis.compute_standard_deviation()
{'x_0': array([0.28892946]), 'x_1': array([0.93716844]), 'x_2': array([0.64592637]), 'x_3': array([0.97884653])}
Get variance¶
Here is the variance for the different variables:
analysis.compute_variance()
{'x_0': array([0.08348023]), 'x_1': array([0.87828468]), 'x_2': array([0.41722087]), 'x_3': array([0.95814053])}
Get quantile¶
Here is the quantile with level 80% for the different variables:
analysis.compute_quantile(0.8)
{'x_0': array([0.79901961]), 'x_1': array([0.72041989]), 'x_2': array([1.44908977]), 'x_3': array([1.5768581])}
Get quartile¶
Here is the second quartile for the different variables:
analysis.compute_quartile(2)
{'x_0': array([0.49875531]), 'x_1': array([-0.06832097]), 'x_2': array([0.90546442]), 'x_3': array([0.67995009])}
Get percentile¶
Here is the 50th percentile for the different variables:
analysis.compute_percentile(50)
{'x_0': array([0.49875531]), 'x_1': array([-0.06832097]), 'x_2': array([0.90546442]), 'x_3': array([0.67995009])}
Get median¶
Here is the median for the different variables:
analysis.compute_median()
{'x_0': array([0.49875531]), 'x_1': array([-0.06832097]), 'x_2': array([0.90546442]), 'x_3': array([0.67995009])}
Get tolerance interval¶
Here is the two-sided tolerance interval with a coverage level equal to 50% with a confidence level of 95% for the different variables:
analysis.compute_tolerance_interval(0.5)
{'x_0': [Bounds(lower=array([0.24854773]), upper=array([0.75453424]))], 'x_1': [Bounds(lower=array([-0.73596335]), upper=array([0.59932142]))], 'x_2': [Bounds(lower=array([0.44530401]), upper=array([1.36562484]))], 'x_3': [Bounds(lower=array([0.24115604]), upper=array([1.32178328]))]}
Get B-value¶
Here is the B-value for the different variables, which is a left-sided tolerance interval with a coverage level equal to 90% with a confidence level of 95%:
analysis.compute_b_value()
{'x_0': array([[0.09841318]]), 'x_1': array([[-1.33841706]]), 'x_2': array([[0.0300737]]), 'x_3': array([[0.09423241]])}
Plot the distribution¶
We can draw the empirical cumulative distribution function and the empirical probability density function:
analysis.plot()
{'x_0': <Figure size 640x320 with 2 Axes>, 'x_1': <Figure size 640x320 with 2 Axes>, 'x_2': <Figure size 640x320 with 2 Axes>, 'x_3': <Figure size 640x320 with 2 Axes>}
Total running time of the script: (0 minutes 1.045 seconds)