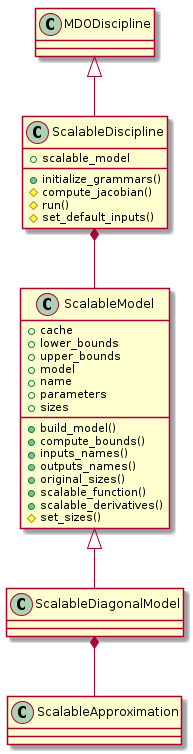
Scalable models¶
Scalability study - API¶
This API facilitates the use of the gemseo.problems.scalable.data_driven.study
package implementing classes to benchmark MDO formulations
based on scalable disciplines.
ScalabilityStudy class implements the concept of scalability study:
By instantiating a
ScalabilityStudy, the user defines the MDO problem in terms of design parameters, objective function and constraints.For each discipline, the user adds a dataset stored in a
Datasetand select a type ofScalableModelto build theScalableDisciplineassociated with this discipline.The user adds different optimization strategies, defined in terms of both optimization algorithms and MDO formulation.
The user adds different scaling strategies, in terms of sizes of design parameters, coupling variables and equality and inequality constraints. The user can also define a scaling strategies according to particular parameters rather than groups of parameters.
Lastly, the user executes the
ScalabilityStudyand the results are written in several files and stored into directories in a hierarchical way, where names depend on both MDO formulation, scaling strategy and replications when it is necessary. Different kinds of files are stored: optimization graphs, dependency matrix plots and of course, scalability results by means of a dedicated class:ScalabilityResult.
- gemseo.problems.scalable.data_driven.api.create_scalability_study(objective, design_variables, directory='study', prefix='', eq_constraints=None, ineq_constraints=None, maximize_objective=False, fill_factor=0.7, active_probability=0.1, feasibility_level=0.8, start_at_equilibrium=True, early_stopping=True, coupling_variables=None)[source]
This method creates a
ScalabilityStudy. It requires two mandatory arguments:the
'objective'name,the list of
'design_variables'names.
Concerning output files, we can specify:
the
directorywhich is'study'by default,the prefix of output file names (default: no prefix).
Regarding optimization parametrization, we can specify:
the list of equality constraints names (
eq_constraints),the list of inequality constraints names (
ineq_constraints),the choice of maximizing the objective function (
maximize_objective).
By default, the objective function is minimized and the MDO problem is unconstrained.
Last but not least, with regard to the scalability methodology, we can overwrite:
the default fill factor of the input-output dependency matrix
ineq_constraints,the probability to set the inequality constraints as active at initial step of the optimization
active_probability,the offset of satisfaction for inequality constraints
feasibility_level,the use of a preliminary MDA to start at equilibrium
start_at_equilibrium,the post-processing of the optimization database to get results earlier than final step
early_stopping.
- Parameters:
objective (str) – name of the objective
design_variables (list(str)) – names of the design variables
directory (str) –
working directory of the study. Default: ‘study’.
By default it is set to “study”.
prefix (str) –
prefix for the output filenames. Default: ‘’.
By default it is set to “”.
eq_constraints (list(str)) – names of the equality constraints. Default: None.
ineq_constraints (list(str)) – names of the inequality constraints Default: None.
maximize_objective (bool) –
maximizing objective. Default: False.
By default it is set to False.
fill_factor (float) –
default fill factor of the input-output dependency matrix. Default: 0.7.
By default it is set to 0.7.
active_probability (float) –
probability to set the inequality constraints as active at initial step of the optimization. Default: 0.1
By default it is set to 0.1.
feasibility_level (float) –
offset of satisfaction for inequality constraints. Default: 0.8.
By default it is set to 0.8.
start_at_equilibrium (bool) –
start at equilibrium using a preliminary MDA. Default: True.
By default it is set to True.
early_stopping (bool) –
post-process the optimization database to get results earlier than final step.
By default it is set to True.
- gemseo.problems.scalable.data_driven.api.plot_scalability_results(study_directory)[source]
This method plots the set of
ScalabilityResultgenerated by aScalabilityStudyand located in the directory created by this study.- Parameters:
study_directory (str) – directory of the scalability study.
Scalable MDO problem¶
This module implements the concept of scalable problem by means of the
ScalableProblem class.
Given
a MDO scenario based on a set of sampled disciplines with a particular problem dimension,
a new problem dimension (= number of inputs and outputs),
a scalable problem:
makes each discipline scalable based on the new problem dimension,
creates the corresponding MDO scenario.
Then, this MDO scenario can be executed and post-processed.
We can repeat this tasks for different sizes of variables and compare the scalability, which is the dependence of the scenario results on the problem dimension.
See also
- class gemseo.problems.scalable.data_driven.problem.ScalableProblem(datasets, design_variables, objective_function, eq_constraints=None, ineq_constraints=None, maximize_objective=False, sizes=None, **parameters)[source]
Scalable problem.
Constructor.
- Parameters:
design_variables (list(str)) – list of design variable names
objective_function (str) – objective function
eq_constraints (list(str)) – equality constraints. Default: None.
eq_constraints – inequality constraints. Default: None.
maximize_objective (bool) –
maximize objective. Default: False.
By default it is set to False.
sizes (dict) – sizes of input and output variables. If None, use the original sizes. Default: None.
parameters – optional parameters for the scalable model.
- create_scenario(formulation='DisciplinaryOpt', scenario_type='MDO', start_at_equilibrium=False, active_probability=0.1, feasibility_level=0.5, **options)[source]
Create a
Scenariofrom the scalable disciplines.- Parameters:
formulation (str) –
The MDO formulation to use for the scenario.
By default it is set to “DisciplinaryOpt”.
scenario_type (str) –
The type of scenario, either
MDOorDOE.By default it is set to “MDO”.
start_at_equilibrium (bool) –
Whether to start at equilibrium using a preliminary MDA.
By default it is set to False.
active_probability (float) –
The probability to set the inequality constraints as active at the initial step of the optimization.
By default it is set to 0.1.
feasibility_level (float) –
The offset of satisfaction for inequality constraints.
By default it is set to 0.5.
**options – The formulation options.
- Returns:
The
Scenariofrom the scalable disciplines.- Return type:
- exec_time(do_sum=True)[source]
Get total execution time per discipline.
- plot_1d_interpolations(save=True, show=False, step=0.01, varnames=None, directory='.', png=False)[source]
Plot 1d interpolations.
- Parameters:
save (bool) –
save plot. Default: True.
By default it is set to True.
show (bool) –
show plot. Default: False.
By default it is set to False.
step (bool) –
Step to evaluate the 1d interpolation function Default: 0.01.
By default it is set to 0.01.
varnames (list(str)) – names of the variable to plot; if None, all variables are plotted. Default: None.
directory (str) –
directory path. Default: ‘.’.
By default it is set to “.”.
png (bool) –
if True, the file format is PNG. Otherwise, use PDF. Default: False.
By default it is set to False.
- plot_coupling_graph()[source]
Plot a coupling graph.
- plot_dependencies(save=True, show=False, directory='.')[source]
Plot dependency matrices.
- plot_n2_chart(save=True, show=False)[source]
Plot a N2 chart.
- property is_feasible
Get the feasibility property of the scenario.
- property n_calls
Get number of disciplinary calls per discipline.
- property n_calls_linearize
Get number of disciplinary calls per discipline.
- property n_calls_linearize_top_level
Get number of top level disciplinary calls per discipline.
- property n_calls_top_level
Get number of top level disciplinary calls per discipline.
- property status
Get the status of the scenario.
Scalable discipline¶
The discipline
implements the concept of scalable discipline.
This is a particular discipline
built from an input-output learning dataset associated with a function
and generalizing its behavior to a new user-defined problem dimension,
that is to say new user-defined input and output dimensions.
Alone or in interaction with other objects of the same type, a scalable discipline can be used to compare the efficiency of an algorithm applying to disciplines with respect to the problem dimension, e.g. optimization algorithm, surrogate model, MDO formulation, MDA, …
The ScalableDiscipline class implements this concept.
It inherits from the MDODiscipline class
in such a way that it can easily be used in a Scenario.
It is composed of a ScalableModel.
The user only needs to provide:
the name of a class overloading
ScalableModel,a dataset as an
Datasetvariables sizes as a dictionary whose keys are the names of inputs and outputs and values are their new sizes. If a variable is missing, its original size is considered.
The ScalableModel parameters can also be filled in,
otherwise the model uses default values.
- class gemseo.problems.scalable.data_driven.discipline.ScalableDiscipline(name, data, sizes=None, **parameters)[source]
Scalable discipline.
Constructor.
- Parameters:
name (str) – scalable model class name.
data (Dataset) – learning dataset.
sizes (dict) – sizes of input and output variables. If None, use the original sizes. Default: None.
parameters – model parameters
name – The name of the discipline. If None, use the class name.
input_grammar_file – The input grammar file path. If
Noneandauto_detect_grammar_files=True, look for"ClassName_input.json"in theGRAMMAR_DIRECTORYif any or in the directory of the discipline class module. IfNoneandauto_detect_grammar_files=False, do not initialize the input grammar from a schema file.output_grammar_file – The output grammar file path. If
Noneandauto_detect_grammar_files=True, look for"ClassName_output.json"in theGRAMMAR_DIRECTORYif any or in the directory of the discipline class module. IfNoneandauto_detect_grammar_files=False, do not initialize the output grammar from a schema file.auto_detect_grammar_files – Whether to look for
"ClassName_{input,output}.json"in theGRAMMAR_DIRECTORYif any or in the directory of the discipline class module when{input,output}_grammar_fileisNone.grammar_type – The type of grammar to define the input and output variables, e.g.
MDODiscipline.JSON_GRAMMAR_TYPEorMDODiscipline.SIMPLE_GRAMMAR_TYPE.cache_type – The type of policy to cache the discipline evaluations, e.g.
MDODiscipline.SIMPLE_CACHEto cache the last one,MDODiscipline.HDF5_CACHEto cache them in an HDF file, orMDODiscipline.MEMORY_FULL_CACHEto cache them in memory. IfNoneor ifactivate_cacheisTrue, do not cache the discipline evaluations.cache_file_path – The HDF file path when
grammar_typeisMDODiscipline.HDF5_CACHE.
- classmethod activate_time_stamps()
Activate the time stamps.
For storing start and end times of execution and linearizations.
- Return type:
None
- add_differentiated_inputs(inputs=None)
Add the inputs against which to differentiate the outputs.
If the discipline grammar type is
MDODiscipline.JSON_GRAMMAR_TYPEand an input is either a non-numeric array or not an array, it will be ignored. If an input is declared as an array but the type of its items is not defined, it is assumed as a numeric array.If the discipline grammar type is
MDODiscipline.SIMPLE_GRAMMAR_TYPEand an input is not an array, it will be ignored. Keep in mind that in this case the array subtype is not checked.- Parameters:
inputs (Iterable[str] | None) – The input variables against which to differentiate the outputs. If None, all the inputs of the discipline are used.
- Raises:
ValueError – When the inputs wrt which differentiate the discipline are not inputs of the latter.
- Return type:
None
- add_differentiated_outputs(outputs=None)
Add the outputs to be differentiated.
If the discipline grammar type is
MDODiscipline.JSON_GRAMMAR_TYPEand an output is either a non-numeric array or not an array, it will be ignored. If an output is declared as an array but the type of its items is not defined, it is assumed as a numeric array.If the discipline grammar type is
MDODiscipline.SIMPLE_GRAMMAR_TYPEand an output is not an array, it will be ignored. Keep in mind that in this case the array subtype is not checked.- Parameters:
outputs (Iterable[str] | None) – The output variables to be differentiated. If None, all the outputs of the discipline are used.
- Raises:
ValueError – When the outputs to differentiate are not discipline outputs.
- Return type:
None
- add_namespace_to_input(name, namespace)
Add a namespace prefix to an existing input grammar element.
The updated input grammar element name will be
namespace+namespaces_separator+name.
- add_namespace_to_output(name, namespace)
Add a namespace prefix to an existing output grammar element.
The updated output grammar element name will be
namespace+namespaces_separator+name.
- add_status_observer(obs)
Add an observer for the status.
Add an observer for the status to be notified when self changes of status.
- Parameters:
obs (Any) – The observer to add.
- Return type:
None
- auto_get_grammar_file(is_input=True, name=None, comp_dir=None)
Use a naming convention to associate a grammar file to the discipline.
Search in the directory
comp_dirfor either an input grammar file namedname + "_input.json"or an output grammar file namedname + "_output.json".- Parameters:
is_input (bool) –
Whether to search for an input or output grammar file.
By default it is set to True.
name (str | None) – The name to be searched in the file names. If
None, use the name of the discipline class.comp_dir (str | Path | None) – The directory in which to search the grammar file. If None, use the
GRAMMAR_DIRECTORYif any, or the directory of the discipline class module.
- Returns:
The grammar file path.
- Return type:
- check_input_data(input_data, raise_exception=True)
Check the input data validity.
- check_jacobian(input_data=None, derr_approx='finite_differences', step=1e-07, threshold=1e-08, linearization_mode='auto', inputs=None, outputs=None, parallel=False, n_processes=2, use_threading=False, wait_time_between_fork=0, auto_set_step=False, plot_result=False, file_path='jacobian_errors.pdf', show=False, fig_size_x=10, fig_size_y=10, reference_jacobian_path=None, save_reference_jacobian=False, indices=None)
Check if the analytical Jacobian is correct with respect to a reference one.
If reference_jacobian_path is not None and save_reference_jacobian is True, compute the reference Jacobian with the approximation method and save it in reference_jacobian_path.
If reference_jacobian_path is not None and save_reference_jacobian is False, do not compute the reference Jacobian but read it from reference_jacobian_path.
If reference_jacobian_path is None, compute the reference Jacobian without saving it.
- Parameters:
input_data (dict[str, ndarray] | None) – The input data needed to execute the discipline according to the discipline input grammar. If None, use the
MDODiscipline.default_inputs.derr_approx (str) –
The approximation method, either “complex_step” or “finite_differences”.
By default it is set to “finite_differences”.
threshold (float) –
The acceptance threshold for the Jacobian error.
By default it is set to 1e-08.
linearization_mode (str) –
the mode of linearization: direct, adjoint or automated switch depending on dimensions of inputs and outputs (Default value = ‘auto’)
By default it is set to “auto”.
inputs (Iterable[str] | None) – The names of the inputs wrt which to differentiate the outputs.
outputs (Iterable[str] | None) – The names of the outputs to be differentiated.
step (float) –
The differentiation step.
By default it is set to 1e-07.
parallel (bool) –
Whether to differentiate the discipline in parallel.
By default it is set to False.
n_processes (int) –
The maximum simultaneous number of threads, if
use_threadingis True, or processes otherwise, used to parallelize the execution.By default it is set to 2.
use_threading (bool) –
Whether to use threads instead of processes to parallelize the execution; multiprocessing will copy (serialize) all the disciplines, while threading will share all the memory This is important to note if you want to execute the same discipline multiple times, you shall use multiprocessing.
By default it is set to False.
wait_time_between_fork (float) –
The time waited between two forks of the process / thread.
By default it is set to 0.
auto_set_step (bool) –
Whether to compute the optimal step for a forward first order finite differences gradient approximation.
By default it is set to False.
plot_result (bool) –
Whether to plot the result of the validation (computed vs approximated Jacobians).
By default it is set to False.
file_path (str | Path) –
The path to the output file if
plot_resultisTrue.By default it is set to “jacobian_errors.pdf”.
show (bool) –
Whether to open the figure.
By default it is set to False.
fig_size_x (float) –
The x-size of the figure in inches.
By default it is set to 10.
fig_size_y (float) –
The y-size of the figure in inches.
By default it is set to 10.
reference_jacobian_path (str | Path | None) – The path of the reference Jacobian file.
save_reference_jacobian (bool) –
Whether to save the reference Jacobian.
By default it is set to False.
indices (Iterable[int] | None) – The indices of the inputs and outputs for the different sub-Jacobian matrices, formatted as
{variable_name: variable_components}wherevariable_componentscan be either an integer, e.g. 2 a sequence of integers, e.g. [0, 3], a slice, e.g. slice(0,3), the ellipsis symbol (…) or None, which is the same as ellipsis. If a variable name is missing, consider all its components. If None, consider all the components of all theinputsandoutputs.
- Returns:
Whether the analytical Jacobian is correct with respect to the reference one.
- check_output_data(raise_exception=True)
Check the output data validity.
- Parameters:
raise_exception (bool) –
Whether to raise an exception when the data is invalid.
By default it is set to True.
- Return type:
None
- classmethod deactivate_time_stamps()
Deactivate the time stamps.
For storing start and end times of execution and linearizations.
- Return type:
None
- static deserialize(file_path)
Deserialize a discipline from a file.
- Parameters:
file_path (str | Path) – The path to the file containing the discipline.
- Returns:
The discipline instance.
- Return type:
- execute(input_data=None)
Execute the discipline.
This method executes the discipline:
Adds the default inputs to the
input_dataif some inputs are not defined in input_data but exist inMDODiscipline.default_inputs.Checks whether the last execution of the discipline was called with identical inputs, i.e. cached in
MDODiscipline.cache; if so, directly returnsself.cache.get_output_cache(inputs).Caches the inputs.
Checks the input data against
MDODiscipline.input_grammar.If
MDODiscipline.data_processoris not None, runs the preprocessor.Updates the status to
MDODiscipline.STATUS_RUNNING.Calls the
MDODiscipline._run()method, that shall be defined.If
MDODiscipline.data_processoris not None, runs the postprocessor.Checks the output data.
Caches the outputs.
Updates the status to
MDODiscipline.STATUS_DONEorMDODiscipline.STATUS_FAILED.Updates summed execution time.
- Parameters:
input_data (Mapping[str, Any] | None) – The input data needed to execute the discipline according to the discipline input grammar. If None, use the
MDODiscipline.default_inputs.- Returns:
The discipline local data after execution.
- Raises:
RuntimeError – When residual_variables are declared but self.run_solves_residuals is False. This is not supported yet.
- Return type:
- get_all_inputs()
Return the local input data as a list.
The order is given by
MDODiscipline.get_input_data_names().
- get_all_outputs()
Return the local output data as a list.
The order is given by
MDODiscipline.get_output_data_names().
- get_attributes_to_serialize()
Define the names of the attributes to be serialized.
Shall be overloaded by disciplines
- static get_data_list_from_dict(keys, data_dict)
Filter the dict from a list of keys or a single key.
If keys is a string, then the method return the value associated to the key. If keys is a list of strings, then the method returns a generator of value corresponding to the keys which can be iterated.
- get_disciplines_in_dataflow_chain()
Return the disciplines that must be shown as blocks in the XDSM.
By default, only the discipline itself is shown. This function can be differently implemented for any type of inherited discipline.
- Returns:
The disciplines shown in the XDSM chain.
- Return type:
- get_expected_dataflow()
Return the expected data exchange sequence.
This method is used for the XDSM representation.
The default expected data exchange sequence is an empty list.
See also
MDOFormulation.get_expected_dataflow
- Returns:
The data exchange arcs.
- Return type:
list[tuple[gemseo.core.discipline.MDODiscipline, gemseo.core.discipline.MDODiscipline, list[str]]]
- get_expected_workflow()
Return the expected execution sequence.
This method is used for the XDSM representation.
The default expected execution sequence is the execution of the discipline itself.
See also
MDOFormulation.get_expected_workflow
- Returns:
The expected execution sequence.
- Return type:
- get_input_data(with_namespaces=True)
Return the local input data as a dictionary.
- get_input_data_names(with_namespaces=True)
Return the names of the input variables.
- get_input_output_data_names(with_namespaces=True)
Return the names of the input and output variables.
- Args:
- with_namespaces: Whether to keep the namespace prefix of the
output names, if any.
- get_inputs_asarray()
Return the local output data as a large NumPy array.
The order is the one of
MDODiscipline.get_all_outputs().- Returns:
The local output data.
- Return type:
- get_inputs_by_name(data_names)
Return the local data associated with input variables.
- Parameters:
data_names (Iterable[str]) – The names of the input variables.
- Returns:
The local data for the given input variables.
- Raises:
ValueError – When a variable is not an input of the discipline.
- Return type:
- get_local_data_by_name(data_names)
Return the local data of the discipline associated with variables names.
- Parameters:
data_names (Iterable[str]) – The names of the variables.
- Returns:
The local data associated with the variables names.
- Raises:
ValueError – When a name is not a discipline input name.
- Return type:
Generator[Any]
- get_output_data(with_namespaces=True)
Return the local output data as a dictionary.
- get_output_data_names(with_namespaces=True)
Return the names of the output variables.
- get_outputs_asarray()
Return the local input data as a large NumPy array.
The order is the one of
MDODiscipline.get_all_inputs().- Returns:
The local input data.
- Return type:
- get_outputs_by_name(data_names)
Return the local data associated with output variables.
- Parameters:
data_names (Iterable[str]) – The names of the output variables.
- Returns:
The local data for the given output variables.
- Raises:
ValueError – When a variable is not an output of the discipline.
- Return type:
- get_sub_disciplines(recursive=False)
Determine the sub-disciplines.
This method lists the sub-disciplines’ disciplines. It will list up to one level of disciplines contained inside another one unless the
recursiveargument is set toTrue.- Parameters:
recursive (bool) –
If
True, the method will look inside any discipline that has other disciplines inside until it reaches a discipline without sub-disciplines, in this case the return value will not include any discipline that has sub-disciplines. IfFalse, the method will list up to one level of disciplines contained inside another one, in this case the return value may include disciplines that contain sub-disciplines.By default it is set to False.
- Returns:
The sub-disciplines.
- Return type:
- initialize_grammars(data)[source]
Initialize input and output grammars from data names.
- Parameters:
data (Dataset) – learning dataset.
- is_all_inputs_existing(data_names)
Test if several variables are discipline inputs.
- is_all_outputs_existing(data_names)
Test if several variables are discipline outputs.
- is_input_existing(data_name)
Test if a variable is a discipline input.
- is_output_existing(data_name)
Test if a variable is a discipline output.
- static is_scenario()
Whether the discipline is a scenario.
- Return type:
- linearize(input_data=None, force_all=False, force_no_exec=False)
Execute the linearized version of the code.
- Parameters:
input_data (Mapping[str, Any] | None) – The input data needed to linearize the discipline according to the discipline input grammar. If None, use the
MDODiscipline.default_inputs.force_all (bool) –
If False,
MDODiscipline._differentiated_inputsandMDODiscipline._differentiated_outputsare used to filter the differentiated variables. otherwise, all outputs are differentiated wrt all inputs.By default it is set to False.
force_no_exec (bool) –
If True, the discipline is not re-executed, cache is loaded anyway.
By default it is set to False.
- Returns:
The Jacobian of the discipline.
- Return type:
- notify_status_observers()
Notify all status observers that the status has changed.
- Return type:
None
- remove_status_observer(obs)
Remove an observer for the status.
- Parameters:
obs (Any) – The observer to remove.
- Return type:
None
- reset_statuses_for_run()
Set all the statuses to
MDODiscipline.STATUS_PENDING.- Raises:
ValueError – When the discipline cannot be run because of its status.
- Return type:
None
- serialize(file_path)
Serialize the discipline and store it in a file.
- Parameters:
file_path (str | Path) – The path to the file to store the discipline.
- Return type:
None
- set_cache_policy(cache_type='SimpleCache', cache_tolerance=0.0, cache_hdf_file=None, cache_hdf_node_name=None, is_memory_shared=True)
Set the type of cache to use and the tolerance level.
This method defines when the output data have to be cached according to the distance between the corresponding input data and the input data already cached for which output data are also cached.
The cache can be either a
SimpleCacherecording the last execution or a cache storing all executions, e.g.MemoryFullCacheandHDF5Cache. Caching data can be either in-memory, e.g.SimpleCacheandMemoryFullCache, or on the disk, e.g.HDF5Cache.The attribute
CacheFactory.cachesprovides the available caches types.- Parameters:
cache_type (str) –
The type of cache.
By default it is set to “SimpleCache”.
cache_tolerance (float) –
The maximum relative norm of the difference between two input arrays to consider that two input arrays are equal.
By default it is set to 0.0.
cache_hdf_file (str | Path | None) – The path to the HDF file to store the data; this argument is mandatory when the
MDODiscipline.HDF5_CACHEpolicy is used.cache_hdf_node_name (str | None) – The name of the HDF file node to store the discipline data. If None,
MDODiscipline.nameis used.is_memory_shared (bool) –
Whether to store the data with a shared memory dictionary, which makes the cache compatible with multiprocessing.
By default it is set to True.
- Return type:
None
- set_disciplines_statuses(status)
Set the sub-disciplines statuses.
To be implemented in subclasses.
- Parameters:
status (str) – The status.
- Return type:
None
- set_jacobian_approximation(jac_approx_type='finite_differences', jax_approx_step=1e-07, jac_approx_n_processes=1, jac_approx_use_threading=False, jac_approx_wait_time=0)
Set the Jacobian approximation method.
Sets the linearization mode to approx_method, sets the parameters of the approximation for further use when calling
MDODiscipline.linearize().- Parameters:
jac_approx_type (str) –
The approximation method, either “complex_step” or “finite_differences”.
By default it is set to “finite_differences”.
jax_approx_step (float) –
The differentiation step.
By default it is set to 1e-07.
jac_approx_n_processes (int) –
The maximum simultaneous number of threads, if
jac_approx_use_threadingis True, or processes otherwise, used to parallelize the execution.By default it is set to 1.
jac_approx_use_threading (bool) –
Whether to use threads instead of processes to parallelize the execution; multiprocessing will copy (serialize) all the disciplines, while threading will share all the memory This is important to note if you want to execute the same discipline multiple times, you shall use multiprocessing.
By default it is set to False.
jac_approx_wait_time (float) –
The time waited between two forks of the process / thread.
By default it is set to 0.
- Return type:
None
- set_optimal_fd_step(outputs=None, inputs=None, force_all=False, print_errors=False, numerical_error=2.220446049250313e-16)
Compute the optimal finite-difference step.
Compute the optimal step for a forward first order finite differences gradient approximation. Requires a first evaluation of the perturbed functions values. The optimal step is reached when the truncation error (cut in the Taylor development), and the numerical cancellation errors (round-off when doing f(x+step)-f(x)) are approximately equal.
Warning
This calls the discipline execution twice per input variables.
See also
https://en.wikipedia.org/wiki/Numerical_differentiation and “Numerical Algorithms and Digital Representation”, Knut Morken , Chapter 11, “Numerical Differentiation”
- Parameters:
inputs (Iterable[str] | None) – The inputs wrt which the outputs are linearized. If None, use the
MDODiscipline._differentiated_inputs.outputs (Iterable[str] | None) – The outputs to be linearized. If None, use the
MDODiscipline._differentiated_outputs.force_all (bool) –
Whether to consider all the inputs and outputs of the discipline;
By default it is set to False.
print_errors (bool) –
Whether to display the estimated errors.
By default it is set to False.
numerical_error (float) –
The numerical error associated to the calculation of f. By default, this is the machine epsilon (appx 1e-16), but can be higher when the calculation of f requires a numerical resolution.
By default it is set to 2.220446049250313e-16.
- Returns:
The estimated errors of truncation and cancellation error.
- Raises:
ValueError – When the Jacobian approximation method has not been set.
- store_local_data(**kwargs)
Store discipline data in local data.
- Parameters:
**kwargs (Any) – The data to be stored in
MDODiscipline.local_data.- Return type:
None
- GRAMMAR_DIRECTORY: ClassVar[str | None] = None
The directory in which to search for the grammar files if not the class one.
- activate_cache: bool = True
Whether to cache the discipline evaluations by default.
- activate_counters: ClassVar[bool] = True
Whether to activate the counters (execution time, calls and linearizations).
- activate_input_data_check: ClassVar[bool] = True
Whether to check the input data respect the input grammar.
- activate_output_data_check: ClassVar[bool] = True
Whether to check the output data respect the output grammar.
- cache: AbstractCache | None
The cache containing one or several executions of the discipline according to the cache policy.
- property cache_tol: float
The cache input tolerance.
This is the tolerance for equality of the inputs in the cache. If norm(stored_input_data-input_data) <= cache_tol * norm(stored_input_data), the cached data for
stored_input_datais returned when callingself.execute(input_data).- Raises:
ValueError – When the discipline does not have a cache.
- data_processor: DataProcessor
A tool to pre- and post-process discipline data.
- property default_inputs: dict[str, Any]
The default inputs.
- Raises:
TypeError – When the default inputs are not passed as a dictionary.
- property disciplines: list[gemseo.core.discipline.MDODiscipline]
The sub-disciplines, if any.
- exec_for_lin: bool
Whether the last execution was due to a linearization.
- property exec_time: float | None
The cumulated execution time of the discipline.
This property is multiprocessing safe.
- Raises:
RuntimeError – When the discipline counters are disabled.
- property grammar_type: str
The type of grammar to be used for inputs and outputs declaration.
- input_grammar: BaseGrammar
The input grammar.
- jac: dict[str, dict[str, ndarray]]
{input:
matrix}}``.
- Type:
The Jacobians of the outputs wrt inputs of the form ``{output
- property linearization_mode: str
The linearization mode among
MDODiscipline.AVAILABLE_MODES.- Raises:
ValueError – When the linearization mode is unknown.
- property local_data: DisciplineData
The current input and output data.
- property n_calls: int | None
The number of times the discipline was executed.
This property is multiprocessing safe.
- Raises:
RuntimeError – When the discipline counters are disabled.
- property n_calls_linearize: int | None
The number of times the discipline was linearized.
This property is multiprocessing safe.
- Raises:
RuntimeError – When the discipline counters are disabled.
- name: str
The name of the discipline.
- output_grammar: BaseGrammar
The output grammar.
- re_exec_policy: str
The policy to re-execute the same discipline.
- residual_variables: Mapping[str, str]
The output variables mapping to their inputs, to be considered as residuals; they shall be equal to zero.
- run_solves_residuals: bool
If True, the run method shall solve the residuals.
- property status: str
The status of the discipline.
The status aims at monitoring the process and give the user a simplified view on the state (the process state = execution or linearize or done) of the disciplines. The core part of the execution is _run, the core part of linearize is _compute_jacobian or approximate jacobian computation.
Scalable model factory¶
This module contains the ScalableModelFactory which is a factory
to create a ScalableModel from its class name by means of the
ScalableModelFactory.create() method. It is also possible to get a list
of available scalable models
(see ScalableModelFactory.scalable_models method)
and to check is a type of scalable model is available
(see ScalableModelFactory.is_available() method)
- class gemseo.problems.scalable.data_driven.factory.ScalableModelFactory[source]
This factory instantiates a class:.ScalableModel from its class name.
The class can be internal to GEMSEO or located in an external module whose path is provided to the constructor.
Initializes the factory: scans the directories to search for subclasses of ScalableModel.
Searches in “GEMSEO_PATH” and gemseo.caches
- create(model_name, data, sizes=None, **parameters)[source]
Create a scalable model.
- is_available(model_name)[source]
Checks the availability of a scalable model.
Scalable model¶
This module implements the abstract concept of scalable model which is used by scalable disciplines. A scalable model is built from an input-output learning dataset associated with a function and generalizing its behavior to a new user-defined problem dimension, that is to say new user-defined input and output dimensions.
The concept of scalable model is implemented
through ScalableModel, an abstract class which is instantiated from:
data provided as a
Datasetvariables sizes provided as a dictionary whose keys are the names of inputs and outputs and values are their new sizes. If a variable is missing, its original size is considered.
Scalable model parameters can also be filled in. Otherwise, the model uses default values.
See also
The ScalableDiagonalModel class overloads ScalableModel.
- class gemseo.problems.scalable.data_driven.model.ScalableModel(data, sizes=None, **parameters)[source]
Scalable model.
Constructor.
- Parameters:
- build_model()[source]
Build model with original sizes for input and output variables.
- compute_bounds()[source]
Compute lower and upper bounds of both input and output variables.
- normalize_data()[source]
Normalize dataset from lower and upper bounds.
- scalable_derivatives(input_value=None)[source]
Evaluate the scalable derivatives.
- scalable_function(input_value=None)[source]
Evaluate the scalable function.
- property original_sizes
Original sizes of variables.
- Returns:
original sizes of variables.
- Return type:
Scalable diagonal model¶
This module implements the concept of scalable diagonal model, which is a particular scalable model built from an input-output dataset relying on a diagonal design of experiments (DOE) where inputs vary proportionally from their lower bounds to their upper bounds, following the diagonal of the input space.
So for every output, the dataset catches its evolution with respect to this proportion, which makes it a mono dimensional behavior. Then, for a new user-defined problem dimension, the scalable model extrapolates this mono dimensional behavior to the different input directions.
The concept of scalable diagonal model is implemented through
the ScalableDiagonalModel class
which is composed of a ScalableDiagonalApproximation.
With regard to the diagonal DOE, GEMSEO proposes the
DiagonalDOE class.
- class gemseo.problems.scalable.data_driven.diagonal.ScalableDiagonalApproximation(sizes, output_dependency, io_dependency, seed=0)[source]
Methodology that captures the trends of a physical problem, and extends it into a problem that has scalable input and outputs dimensions The original and the resulting scalable problem have the same interface:
all inputs and outputs have the same names; only their dimensions vary.
Constructor:
- Parameters:
- build_scalable_function(function_name, dataset, input_names, degree=3)[source]
Build interpolation from a 1D input and output function. Add the model to the local dictionary.
- get_scalable_derivative(output_function)[source]
Retrieve the (scalable) gradient of the scalable function generated from the original discipline.
- Parameters:
output_function (str) – name of the output function
- class gemseo.problems.scalable.data_driven.diagonal.ScalableDiagonalModel(data, sizes=None, fill_factor=-1, comp_dep=None, inpt_dep=None, force_input_dependency=False, allow_unused_inputs=True, seed=1, group_dep=None)[source]
Scalable diagonal model.
Constructor.
- Parameters:
data (Dataset) – learning dataset.
sizes (dict) – sizes of input and output variables. If None, use the original sizes. Default: None.
fill_factor –
degree of sparsity of the dependency matrix. Default: -1.
By default it is set to -1.
comp_dep – matrix that establishes the selection of a single original component for each scalable component
inpt_dep – dependency matrix that establishes the dependency of outputs wrt inputs
force_input_dependency (bool) –
for any output, force dependency with at least on input.
By default it is set to False.
allow_unused_inputs (bool) –
possibility to have an input with no dependence with any output
By default it is set to True.
seed (int) –
seed
By default it is set to 1.
group_dep (dict(list(str))) – dependency between inputs and outputs
- build_model()[source]
Build model with original sizes for input and output variables.
- Returns:
scalable approximation.
- Return type:
- compute_bounds()
Compute lower and upper bounds of both input and output variables.
- generate_random_dependency()[source]
Generates a random dependency structure for use in scalable discipline.
- normalize_data()
Normalize dataset from lower and upper bounds.
- plot_1d_interpolations(save=False, show=False, step=0.01, varnames=None, directory='.', png=False)[source]
Plot the scaled 1D interpolations, a.k.a. the basis functions.
A basis function is a mono dimensional function interpolating the samples of a given output component over the input sampling line \(t\in[0,1]\mapsto \\underline{x}+t(\overline{x}-\\underline{x})\).
There are as many basis functions as there are output components from the discipline. Thus, for a discipline with a single output in dimension 1, there is 1 basis function. For a discipline with a single output in dimension 2, there are 2 basis functions. For a discipline with an output in dimension 2 and an output in dimension 13, there are 15 basis functions. And so on. This method allows to plot the basis functions associated with all outputs or only part of them, either on screen (
show=True), in a file (save=True) or both. We can also specify the discretizationstepwhose default value is0.01.- Parameters:
save (bool) –
if True, export the plot as a PDF file (Default value = False)
By default it is set to False.
show (bool) –
if True, display the plot (Default value = False)
By default it is set to False.
step (bool) –
Step to evaluate the 1d interpolation function (Default value = 0.01)
By default it is set to 0.01.
varnames (list(str)) – names of the variable to plot; if None, all variables are plotted (Default value = None)
directory (str) –
directory path. Default: ‘.’.
By default it is set to “.”.
png (bool) –
if True, the file format is PNG. Otherwise, use PDF. Default: False.
By default it is set to False.
- plot_dependency(add_levels=True, save=True, show=False, directory='.', png=False)[source]
This method plots the dependency matrix of a discipline in the form of a chessboard, where rows represent inputs, columns represent output and gray scale represent the dependency level between inputs and outputs.
- Parameters:
add_levels (bool) –
add values of dependency levels in percentage. Default: True.
By default it is set to True.
save (bool) –
if True, export the plot into a file. Default: True.
By default it is set to True.
show (bool) –
if True, display the plot. Default: False.
By default it is set to False.
directory (str) –
directory path. Default: ‘.’.
By default it is set to “.”.
png (bool) –
if True, the file format is PNG. Otherwise, use PDF. Default: False.
By default it is set to False.
- scalable_derivatives(input_value=None)[source]
Evaluate the scalable derivatives.
- scalable_function(input_value=None)[source]
Evaluate the scalable functions.
- property original_sizes
Original sizes of variables.
- Returns:
original sizes of variables.
- Return type:
- gemseo.problems.scalable.data_driven.diagonal.choice(a, size=None, replace=True, p=None)
Generates a random sample from a given 1-D array
New in version 1.7.0.
Note
New code should use the
choicemethod of adefault_rng()instance instead; please see the Quick Start.- Parameters:
a (1-D array-like or int) – If an ndarray, a random sample is generated from its elements. If an int, the random sample is generated as if it were
np.arange(a)size (int or tuple of ints, optional) – Output shape. If the given shape is, e.g.,
(m, n, k), thenm * n * ksamples are drawn. Default is None, in which case a single value is returned.replace (boolean, optional) – Whether the sample is with or without replacement. Default is True, meaning that a value of
acan be selected multiple times.p (1-D array-like, optional) – The probabilities associated with each entry in a. If not given, the sample assumes a uniform distribution over all entries in
a.
- Returns:
samples – The generated random samples
- Return type:
single item or ndarray
- Raises:
ValueError – If a is an int and less than zero, if a or p are not 1-dimensional, if a is an array-like of size 0, if p is not a vector of probabilities, if a and p have different lengths, or if replace=False and the sample size is greater than the population size
Notes
Setting user-specified probabilities through
puses a more general but less efficient sampler than the default. The general sampler produces a different sample than the optimized sampler even if each element ofpis 1 / len(a).Sampling random rows from a 2-D array is not possible with this function, but is possible with Generator.choice through its
axiskeyword.Examples
Generate a uniform random sample from np.arange(5) of size 3:
>>> np.random.choice(5, 3) array([0, 3, 4]) # random >>> #This is equivalent to np.random.randint(0,5,3)
Generate a non-uniform random sample from np.arange(5) of size 3:
>>> np.random.choice(5, 3, p=[0.1, 0, 0.3, 0.6, 0]) array([3, 3, 0]) # random
Generate a uniform random sample from np.arange(5) of size 3 without replacement:
>>> np.random.choice(5, 3, replace=False) array([3,1,0]) # random >>> #This is equivalent to np.random.permutation(np.arange(5))[:3]
Generate a non-uniform random sample from np.arange(5) of size 3 without replacement:
>>> np.random.choice(5, 3, replace=False, p=[0.1, 0, 0.3, 0.6, 0]) array([2, 3, 0]) # random
Any of the above can be repeated with an arbitrary array-like instead of just integers. For instance:
>>> aa_milne_arr = ['pooh', 'rabbit', 'piglet', 'Christopher'] >>> np.random.choice(aa_milne_arr, 5, p=[0.5, 0.1, 0.1, 0.3]) array(['pooh', 'pooh', 'pooh', 'Christopher', 'piglet'], # random dtype='<U11')
- gemseo.problems.scalable.data_driven.diagonal.npseed()
seed(self, seed=None)
Reseed a legacy MT19937 BitGenerator
Notes
This is a convenience, legacy function.
The best practice is to not reseed a BitGenerator, rather to recreate a new one. This method is here for legacy reasons. This example demonstrates best practice.
>>> from numpy.random import MT19937 >>> from numpy.random import RandomState, SeedSequence >>> rs = RandomState(MT19937(SeedSequence(123456789))) # Later, you want to restart the stream >>> rs = RandomState(MT19937(SeedSequence(987654321)))
- gemseo.problems.scalable.data_driven.diagonal.rand(d0, d1, ..., dn)
Random values in a given shape.
Note
This is a convenience function for users porting code from Matlab, and wraps random_sample. That function takes a tuple to specify the size of the output, which is consistent with other NumPy functions like numpy.zeros and numpy.ones.
Create an array of the given shape and populate it with random samples from a uniform distribution over
[0, 1).- Parameters:
d0 (int, optional) – The dimensions of the returned array, must be non-negative. If no argument is given a single Python float is returned.
d1 (int, optional) – The dimensions of the returned array, must be non-negative. If no argument is given a single Python float is returned.
... (int, optional) – The dimensions of the returned array, must be non-negative. If no argument is given a single Python float is returned.
dn (int, optional) – The dimensions of the returned array, must be non-negative. If no argument is given a single Python float is returned.
- Returns:
out – Random values.
- Return type:
ndarray, shape
(d0, d1, ..., dn)
See also
Examples
>>> np.random.rand(3,2) array([[ 0.14022471, 0.96360618], #random [ 0.37601032, 0.25528411], #random [ 0.49313049, 0.94909878]]) #random
- gemseo.problems.scalable.data_driven.diagonal.randint(low, high=None, size=None, dtype=int)
Return random integers from low (inclusive) to high (exclusive).
Return random integers from the “discrete uniform” distribution of the specified dtype in the “half-open” interval [low, high). If high is None (the default), then results are from [0, low).
Note
New code should use the
integersmethod of adefault_rng()instance instead; please see the Quick Start.- Parameters:
low (int or array-like of ints) – Lowest (signed) integers to be drawn from the distribution (unless
high=None, in which case this parameter is one above the highest such integer).high (int or array-like of ints, optional) – If provided, one above the largest (signed) integer to be drawn from the distribution (see above for behavior if
high=None). If array-like, must contain integer valuessize (int or tuple of ints, optional) – Output shape. If the given shape is, e.g.,
(m, n, k), thenm * n * ksamples are drawn. Default is None, in which case a single value is returned.dtype (dtype, optional) –
Desired dtype of the result. Byteorder must be native. The default value is int.
New in version 1.11.0.
- Returns:
out – size-shaped array of random integers from the appropriate distribution, or a single such random int if size not provided.
- Return type:
int or ndarray of ints
See also
random_integerssimilar to randint, only for the closed interval [low, high], and 1 is the lowest value if high is omitted.
random.Generator.integerswhich should be used for new code.
Examples
>>> np.random.randint(2, size=10) array([1, 0, 0, 0, 1, 1, 0, 0, 1, 0]) # random >>> np.random.randint(1, size=10) array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0])
Generate a 2 x 4 array of ints between 0 and 4, inclusive:
>>> np.random.randint(5, size=(2, 4)) array([[4, 0, 2, 1], # random [3, 2, 2, 0]])
Generate a 1 x 3 array with 3 different upper bounds
>>> np.random.randint(1, [3, 5, 10]) array([2, 2, 9]) # random
Generate a 1 by 3 array with 3 different lower bounds
>>> np.random.randint([1, 5, 7], 10) array([9, 8, 7]) # random
Generate a 2 by 4 array using broadcasting with dtype of uint8
>>> np.random.randint([1, 3, 5, 7], [[10], [20]], dtype=np.uint8) array([[ 8, 6, 9, 7], # random [ 1, 16, 9, 12]], dtype=uint8)