Note
Click here to download the full example code
Scalable diagonal discipline¶
Let us consider the
SobieskiAerodynamics discipline.
We want to build its ScalableDiscipline counterpart,
using a ScalableDiagonalModel
For that, we can use a 20-length DiagonalDOE
and test different sizes of variables or different settings
for the scalable diagonal discipline.
from __future__ import division, unicode_literals
from matplotlib import pyplot as plt
Import¶
from gemseo.api import (
configure_logger,
create_discipline,
create_scalable,
create_scenario,
)
from gemseo.problems.sobieski.core import SobieskiProblem
configure_logger()
Out:
<RootLogger root (INFO)>
Learning dataset¶
The first step is to build an AbstractFullCache dataset
from a DiagonalDOE.
Instantiate the discipline¶
For that, we instantiate the
SobieskiAerodynamics discipline
and set it up to cache all evaluations.
discipline = create_discipline("SobieskiAerodynamics")
discipline.set_cache_policy(discipline.MEMORY_FULL_CACHE)
Get the input space¶
We also define the input space on which to sample the discipline.
input_space = SobieskiProblem().read_design_space()
input_space.filter(discipline.get_input_data_names())
Out:
<gemseo.algos.design_space.DesignSpace object at 0x7fcaa46148e0>
Build the DOE scenario¶
Lastly, we sample the discipline by means of a DOEScenario
relying on both discipline and input space.
In order to build a diagonal scalable discipline,
a DiagonalDOE must be used.
scenario = create_scenario(
[discipline], "DisciplinaryOpt", "y_2", input_space, scenario_type="DOE"
)
scenario.execute({"algo": "DiagonalDOE", "n_samples": 20})
Out:
INFO - 14:42:51:
INFO - 14:42:51: *** Start DOE Scenario execution ***
INFO - 14:42:51: DOEScenario
INFO - 14:42:51: Disciplines: SobieskiAerodynamics
INFO - 14:42:51: MDOFormulation: DisciplinaryOpt
INFO - 14:42:51: Algorithm: DiagonalDOE
INFO - 14:42:51: Optimization problem:
INFO - 14:42:51: Minimize: y_2(x_shared, x_2, y_32, y_12)
INFO - 14:42:51: With respect to: x_shared, x_2, y_32, y_12
INFO - 14:42:51: DOE sampling: 0%| | 0/20 [00:00<?, ?it]
INFO - 14:42:51: DOE sampling: 100%|██████████| 20/20 [00:00<00:00, 437.83 it/sec, obj=[7.72500000e+04 1.27392070e+04 6.06395673e+00]]
INFO - 14:42:51: Optimization result:
INFO - 14:42:51: Objective value = 25508.372961119574
INFO - 14:42:51: The result is feasible.
INFO - 14:42:51: Status: None
INFO - 14:42:51: Optimizer message: None
INFO - 14:42:51: Number of calls to the objective function by the optimizer: 20
INFO - 14:42:51: Design space:
INFO - 14:42:51: +----------+-------------+-------+-------------+-------+
INFO - 14:42:51: | name | lower_bound | value | upper_bound | type |
INFO - 14:42:51: +----------+-------------+-------+-------------+-------+
INFO - 14:42:51: | x_shared | 0.01 | 0.01 | 0.09 | float |
INFO - 14:42:51: | x_shared | 30000 | 30000 | 60000 | float |
INFO - 14:42:51: | x_shared | 1.4 | 1.4 | 1.8 | float |
INFO - 14:42:51: | x_shared | 2.5 | 2.5 | 8.5 | float |
INFO - 14:42:51: | x_shared | 40 | 40 | 70 | float |
INFO - 14:42:51: | x_shared | 500 | 500 | 1500 | float |
INFO - 14:42:51: | x_2 | 0.75 | 0.75 | 1.25 | float |
INFO - 14:42:51: | y_32 | 0.235 | 0.235 | 0.795 | float |
INFO - 14:42:51: | y_12 | 24850 | 24850 | 77250 | float |
INFO - 14:42:51: | y_12 | 0.45 | 0.45 | 1.5 | float |
INFO - 14:42:51: +----------+-------------+-------+-------------+-------+
INFO - 14:42:51: *** DOE Scenario run terminated ***
{'eval_jac': False, 'algo': 'DiagonalDOE', 'n_samples': 20}
Scalable diagonal discipline¶
Build the scalable discipline¶
The second step is to build a ScalableDiscipline,
using a ScalableDiagonalModel and the cache of the discipline,
converted to a Dataset.
dataset = discipline.cache.export_to_dataset()
scalable = create_scalable("ScalableDiagonalModel", dataset)
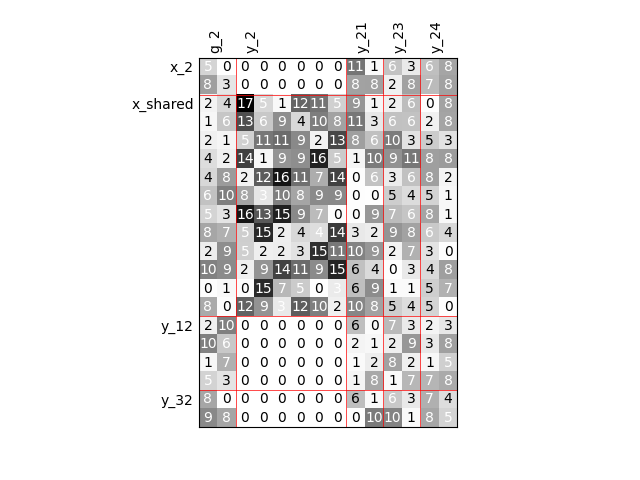
Visualize the input-output dependencies¶
We can easily access the underlying ScalableDiagonalModel
and plot the corresponding input-output dependency matrix
where the level of gray and the number (in [0,100]) represent
the degree of dependency between inputs and outputs.
Input are on the left while outputs are at the top.
More precisely, for a given output component located at the top of the graph,
these degrees are contributions to the output component and they add up to 1.
In other words, a degree expresses this contribution in percentage
and for a given column, the elements add up to 100.
scalable.scalable_model.plot_dependency(save=False, show=False)
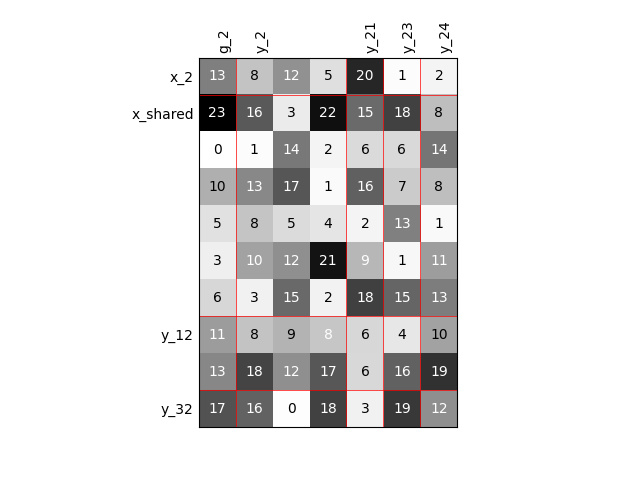
Out:
'None'
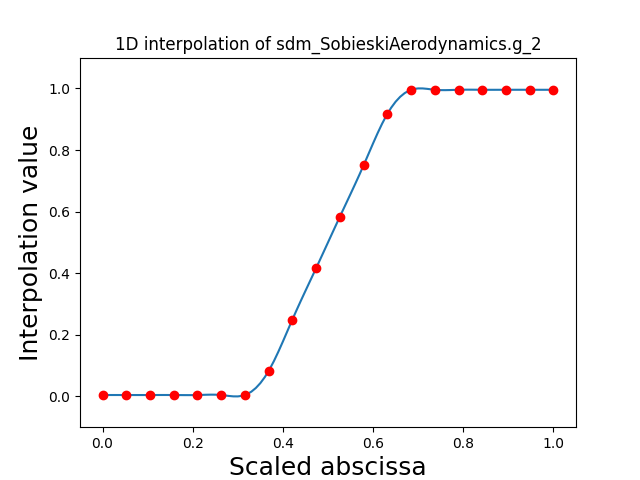
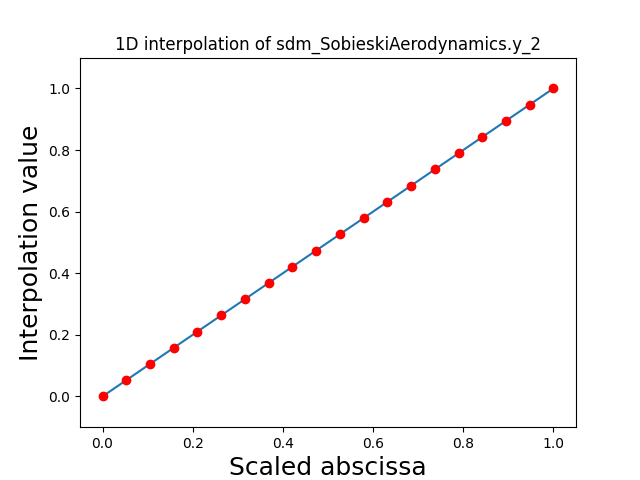
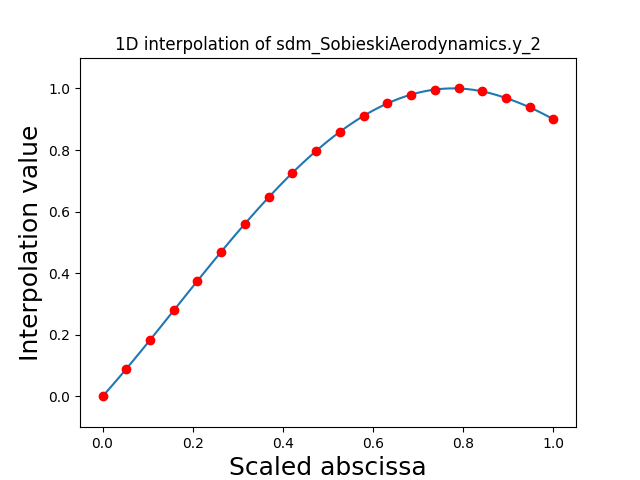
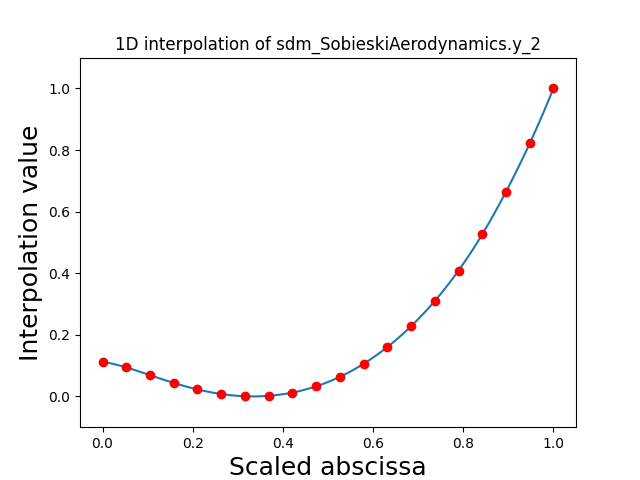
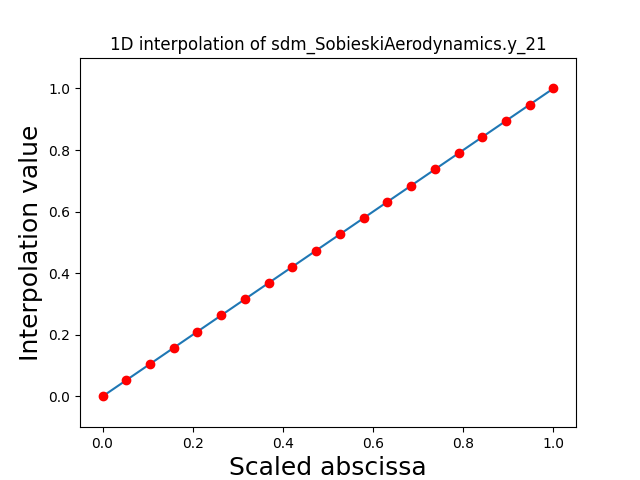
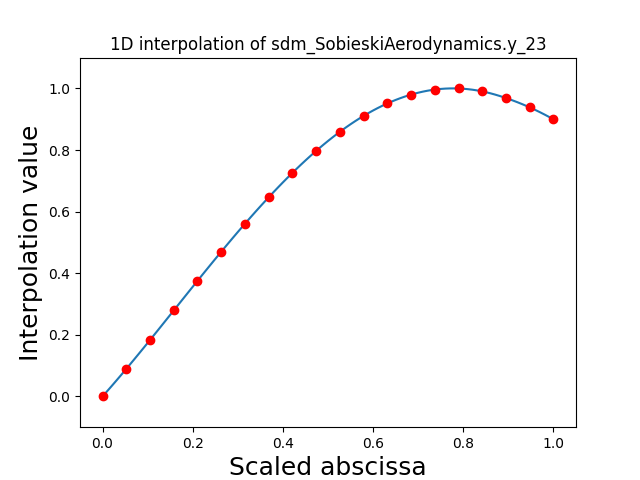
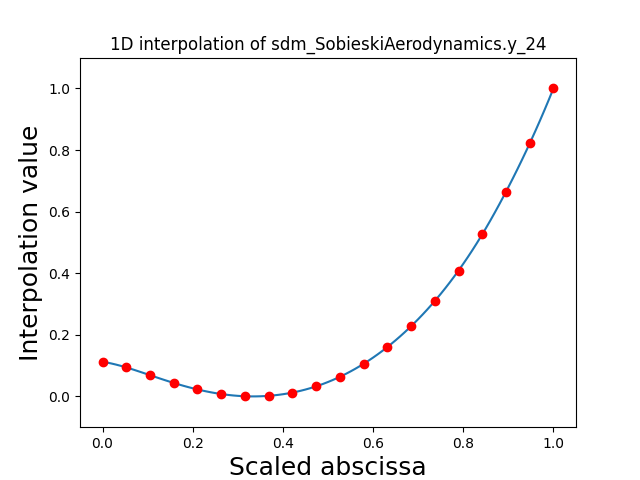
Visualize the 1D interpolations¶
For every output, we can also visualize a spline interpolation of the output samples over the diagonal of the input space.
scalable.scalable_model.plot_1d_interpolations(save=False, show=False)
Out:
[]
Increased problem dimension¶
We can repeat the construction of the scalable discipline for different sizes of variables and visualize the input-output dependency matrices.
Twice as many inputs¶
For example, we can increase the size of each input by a factor of 2.
sizes = {
name: discipline.cache.varsizes[name] * 2
for name in discipline.get_input_data_names()
}
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes)
scalable.scalable_model.plot_dependency(save=False, show=False)
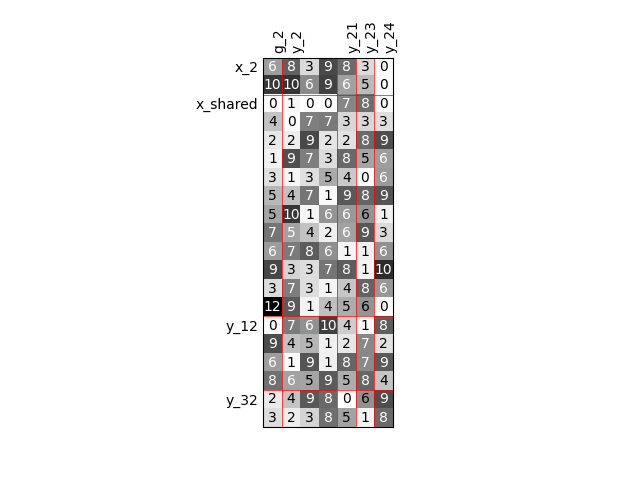
Out:
'None'
Twice as many outputs¶
Or we can increase the size of each output by a factor of 2.
sizes = {
name: discipline.cache.varsizes[name] * 2
for name in discipline.get_output_data_names()
}
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes)
scalable.scalable_model.plot_dependency(save=False, show=False)
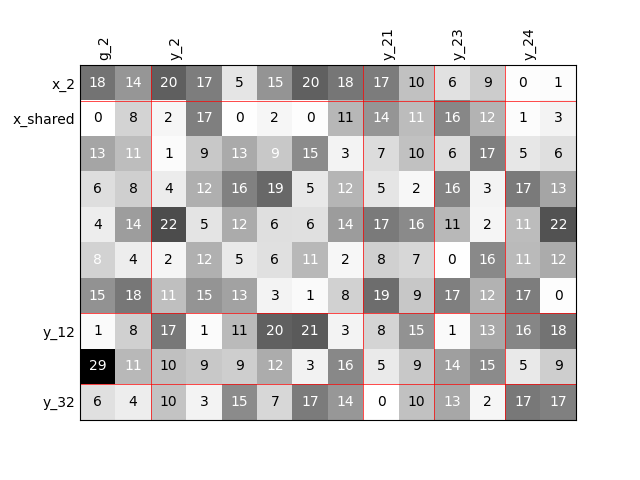
Out:
'None'
Twice as many variables¶
Or we can increase the size of each input and each output by a factor of 2.
names = list(discipline.get_input_data_names())
names += list(discipline.get_output_data_names())
sizes = {name: discipline.cache.varsizes[name] * 2 for name in names}
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes)
scalable.scalable_model.plot_dependency(save=False, show=False)
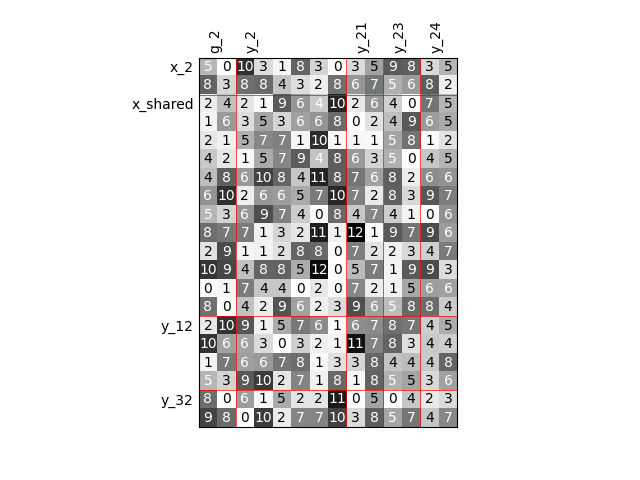
Out:
'None'
Binary IO dependencies¶
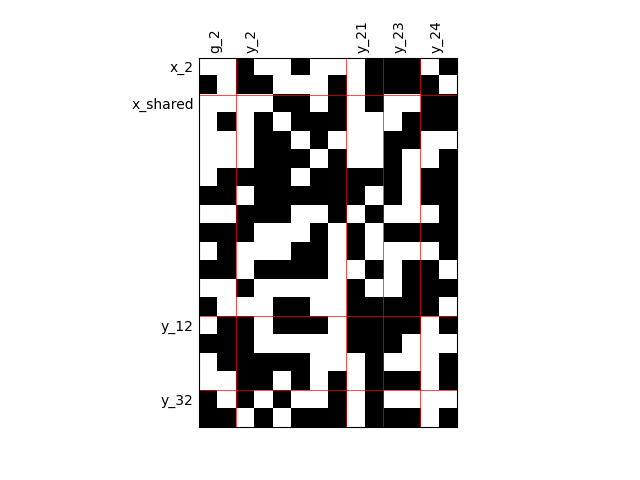
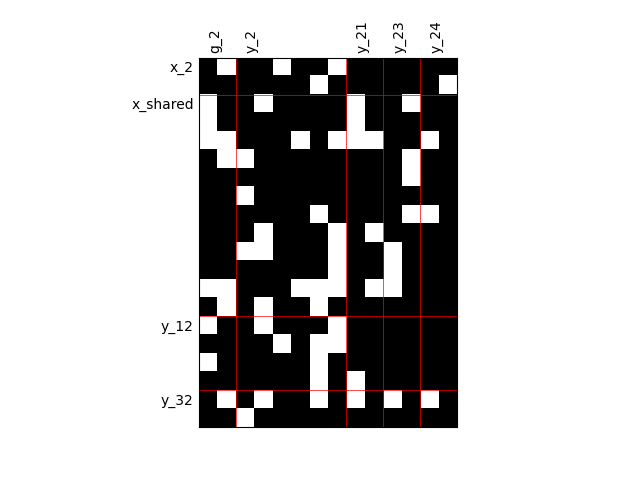
By default, any output component depends on any input component with a random level. We can also consider sparser input-output dependency by means of binary input-output dependency matrices. For that, we have to set the value of the fill factor which represents the part of connection between inputs and outputs. Then, a connection is represented by a black square while an absence of connection is presented by a white one. When the fill factor is equal to 1, any input is connected to any output. Conversely, when the fill factor is equal to 0, there is not a single connection between inputs and outputs.
Fill factor = 0.2¶
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes, fill_factor=0.2)
scalable.scalable_model.plot_dependency(save=False, show=False)
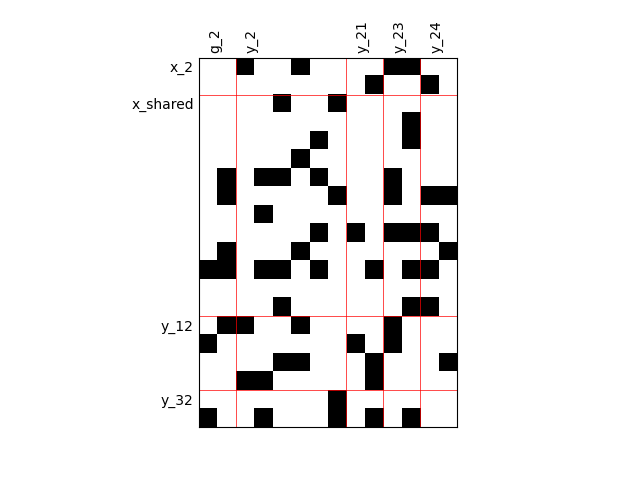
Out:
'None'
Fill factor = 0.5¶
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes, fill_factor=0.5)
scalable.scalable_model.plot_dependency(save=False, show=False)
Out:
'None'
Fill factor = 0.8¶
scalable = create_scalable("ScalableDiagonalModel", dataset, sizes, fill_factor=0.8)
scalable.scalable_model.plot_dependency(save=False, show=False)
Out:
'None'
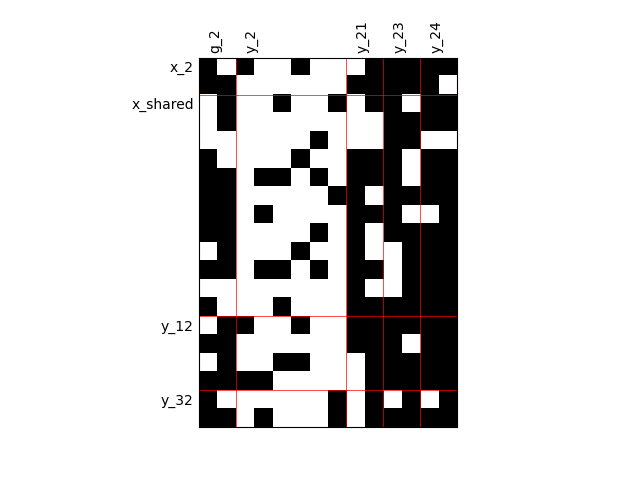
Heterogeneous dependencies¶
scalable = create_scalable(
"ScalableDiagonalModel", dataset, sizes, fill_factor={"y_2": 0.2}
)
scalable.scalable_model.plot_dependency(save=False, show=False)
Out:
'None'
Group dependencies¶
scalable = create_scalable(
"ScalableDiagonalModel", dataset, sizes, group_dep={"y_2": ["x_shared"]}
)
scalable.scalable_model.plot_dependency(save=False, show=False)
# Workaround for HTML rendering, instead of ``show=True``
plt.show()
Total running time of the script: ( 0 minutes 3.235 seconds)