Note
Go to the end to download the full example code
Morris analysis¶
from __future__ import annotations
import pprint
from gemseo.uncertainty.sensitivity.morris.analysis import MorrisAnalysis
from gemseo.uncertainty.use_cases.ishigami.ishigami_discipline import IshigamiDiscipline
from gemseo.uncertainty.use_cases.ishigami.ishigami_space import IshigamiSpace
In this example, we consider the Ishigami function [IH90]
implemented as an MDODiscipline by the IshigamiDiscipline.
It is commonly used
with the independent random variables \(X_1\), \(X_2\) and \(X_3\)
uniformly distributed between \(-\pi\) and \(\pi\)
and defined in the IshigamiSpace.
discipline = IshigamiDiscipline()
uncertain_space = IshigamiSpace()
Then,
we run sensitivity analysis of type MorrisAnalysis:
sensitivity_analysis = MorrisAnalysis([discipline], uncertain_space, 10)
sensitivity_analysis.compute_indices()
{'MU': {'y': [{'x1': array([0.73532408]), 'x2': array([-0.05115399]), 'x3': array([-1.6024484])}]}, 'MU_STAR': {'y': [{'x1': array([0.76770333]), 'x2': array([2.09435091]), 'x3': array([1.6024484])}]}, 'SIGMA': {'y': [{'x1': array([0.76770333]), 'x2': array([2.09435091]), 'x3': array([1.58984353])}]}, 'RELATIVE_SIGMA': {'y': [{'x1': array([1.]), 'x2': array([1.]), 'x3': array([0.99213399])}]}, 'MIN': {'y': [{'x1': array([0.03237925]), 'x2': array([2.04319692]), 'x3': array([0.01260487])}]}, 'MAX': {'y': [{'x1': array([1.50302741]), 'x2': array([2.14550491]), 'x3': array([3.19229192])}]}}
The resulting indices are the empirical means and the standard deviations of the absolute output variations due to input changes.
pprint.pprint(sensitivity_analysis.indices)
{'MAX': {'y': [{'x1': array([1.50302741]),
'x2': array([2.14550491]),
'x3': array([3.19229192])}]},
'MIN': {'y': [{'x1': array([0.03237925]),
'x2': array([2.04319692]),
'x3': array([0.01260487])}]},
'MU': {'y': [{'x1': array([0.73532408]),
'x2': array([-0.05115399]),
'x3': array([-1.6024484])}]},
'MU_STAR': {'y': [{'x1': array([0.76770333]),
'x2': array([2.09435091]),
'x3': array([1.6024484])}]},
'RELATIVE_SIGMA': {'y': [{'x1': array([1.]),
'x2': array([1.]),
'x3': array([0.99213399])}]},
'SIGMA': {'y': [{'x1': array([0.76770333]),
'x2': array([2.09435091]),
'x3': array([1.58984353])}]}}
The main indices corresponds to these empirical means
(this main method can be changed with MorrisAnalysis.main_method):
pprint.pprint(sensitivity_analysis.main_indices)
{'y': [{'x1': array([0.76770333]),
'x2': array([2.09435091]),
'x3': array([1.6024484])}]}
and can be interpreted with respect to the empirical bounds of the outputs:
pprint.pprint(sensitivity_analysis.outputs_bounds)
{'y': [array([0.3861887]), array([14.38383568])]}
We can also get the input parameters sorted by decreasing order of influence:
sensitivity_analysis.sort_parameters("y")
['x2', 'x3', 'x1']
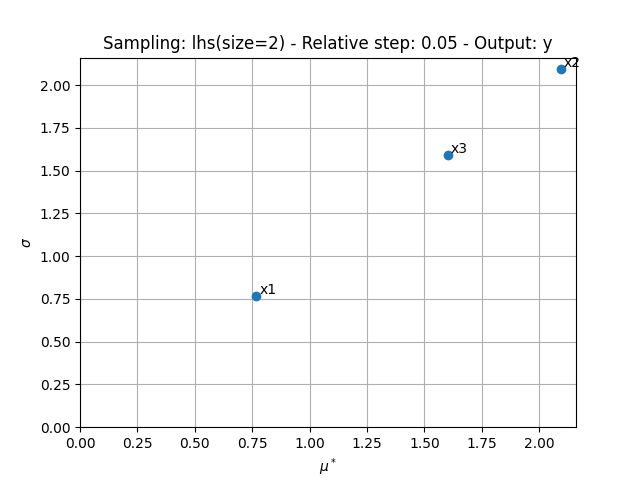
We can use the method MorrisAnalysis.plot()
to visualize the different series of indices:
sensitivity_analysis.plot("y", save=False, show=True, lower_mu=0, lower_sigma=0)
Lastly,
the sensitivity indices can be exported to a Dataset:
sensitivity_analysis.to_dataset()
Total running time of the script: (0 minutes 0.477 seconds)
