Note
Go to the end to download the full example code
R2 for regression models¶
from matplotlib import pyplot as plt
from numpy import array
from numpy import linspace
from numpy import newaxis
from numpy import sin
from gemseo.datasets.io_dataset import IODataset
from gemseo.mlearning.quality_measures.r2_measure import R2Measure
from gemseo.mlearning.regression.polyreg import PolynomialRegressor
from gemseo.mlearning.regression.rbf import RBFRegressor
Given a dataset \((x_i,y_i,\hat{y}_i)_{1\leq i \leq N}\) where \(x_i\) is an input point, \(y_i\) is an output observation and \(\hat{y}_i=\hat{f}(x_i)\) is an output prediction computed by a regression model \(\hat{f}\), the \(R^2\) metric (also known as \(Q^2\)) is written
where \(\bar{y}=\frac{1}{N}\sum_{i=1}^Ny_i\). The higher, the better. From 0.9 it starts to look (very) good. A negative value is very bad; a constant model would do better.
To illustrate this quality measure, let us consider the function \(f(x)=(6x-2)^2\sin(12x-4)\) [FSK08]:
def f(x):
return (6 * x - 2) ** 2 * sin(12 * x - 4)
and try to approximate it with a polynomial of order 3.
For this, we can take these 7 learning input points
x_train = array([0.1, 0.3, 0.5, 0.6, 0.8, 0.9, 0.95])
and evaluate the model f over this design of experiments (DOE):
y_train = f(x_train)
Then,
we create an IODataset from these 7 learning samples:
dataset_train = IODataset()
dataset_train.add_input_group(x_train[:, newaxis], ["x"])
dataset_train.add_output_group(y_train[:, newaxis], ["y"])
and build a PolynomialRegressor with degree=3 from it:
polynomial = PolynomialRegressor(dataset_train, 3)
polynomial.learn()
Before using it, we are going to measure its quality with the \(R^2\) metric:
r2 = R2Measure(polynomial)
r2.compute_learning_measure()
array([0.78649338])
This result is medium, and we can be expected to a poor generalization quality. As the cost of this academic function is zero, we can approximate this generalization quality with a large test dataset whereas the usual test size is about 20% of the training size.
x_test = linspace(0.0, 1.0, 100)
y_test = f(x_test)
dataset_test = IODataset()
dataset_test.add_input_group(x_test[:, newaxis], ["x"])
dataset_test.add_output_group(y_test[:, newaxis], ["y"])
r2.compute_test_measure(dataset_test)
array([0.47280012])
The quality is lower than 0.5, which is pretty mediocre. This can be explained by a broader generalization domain than that of learning, which highlights the difficulties of extrapolation:
plt.plot(x_test, y_test, "-b", label="Reference")
plt.plot(x_train, y_train, "ob")
plt.plot(x_test, polynomial.predict(x_test[:, newaxis]), "-r", label="Prediction")
plt.plot(x_train, polynomial.predict(x_train[:, newaxis]), "or")
plt.legend()
plt.grid()
plt.show()
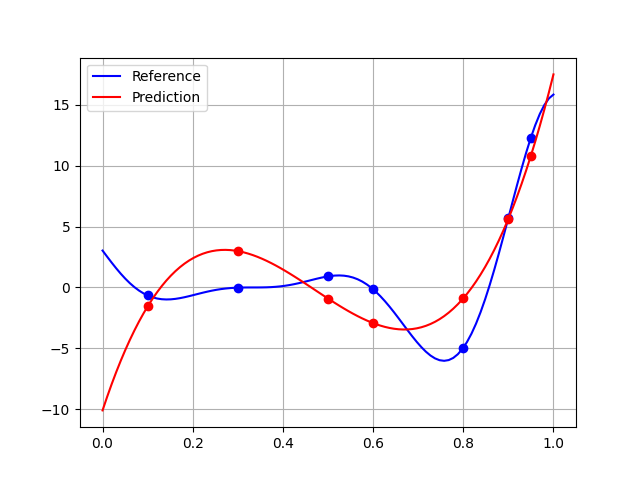
Using the learning domain would slightly improve the quality:
x_test = linspace(x_train.min(), x_train.max(), 100)
y_test = f(x_test)
dataset_test_in_learning_domain = IODataset()
dataset_test_in_learning_domain.add_input_group(x_test[:, newaxis], ["x"])
dataset_test_in_learning_domain.add_output_group(y_test[:, newaxis], ["y"])
r2.compute_test_measure(dataset_test_in_learning_domain)
array([0.50185268])
Lastly, to get better results without new learning points, we would have to change the regression model:
rbf = RBFRegressor(dataset_train)
rbf.learn()
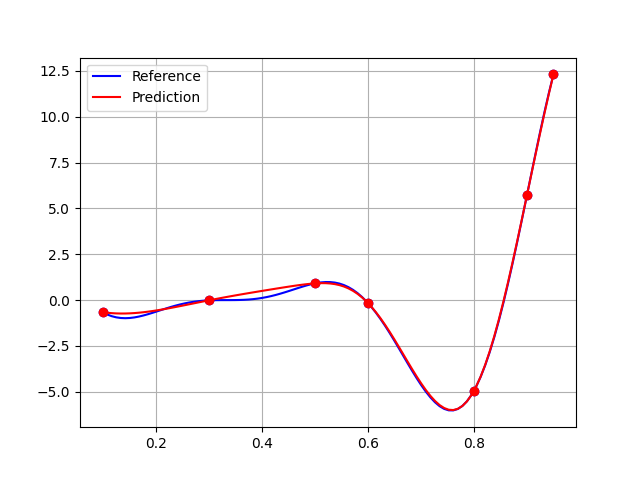
The quality of this RBFRegressor is quite good,
both on the learning side:
r2_rbf = R2Measure(rbf)
r2_rbf.compute_learning_measure()
array([1.])
and on the validation side:
r2_rbf.compute_test_measure(dataset_test_in_learning_domain)
array([0.99807284])
including the larger domain:
r2_rbf.compute_test_measure(dataset_test)
array([0.98593573])
A final plot to convince us:
plt.plot(x_test, y_test, "-b", label="Reference")
plt.plot(x_train, y_train, "ob")
plt.plot(x_test, rbf.predict(x_test[:, newaxis]), "-r", label="Prediction")
plt.plot(x_train, rbf.predict(x_train[:, newaxis]), "or")
plt.legend()
plt.grid()
plt.show()
Total running time of the script: (0 minutes 0.315 seconds)
