Note
Click here to download the full example code
Iris dataset¶
Presentation¶
This is one of the best known dataset to be found in the machine learning literature.
It was introduced by the statistician Ronald Fisher in his 1936 paper “The use of multiple measurements in taxonomic problems”, Annals of Eugenics. 7 (2): 179–188.
It contains 150 instances of iris plants:
50 Iris Setosa,
50 Iris Versicolour,
50 Iris Virginica.
Each instance is characterized by:
its sepal length in cm,
its sepal width in cm,
its petal length in cm,
its petal width in cm.
This dataset can be used for either clustering purposes or classification ones.
from __future__ import annotations
from gemseo.api import configure_logger
from gemseo.api import load_dataset
from gemseo.post.dataset.andrews_curves import AndrewsCurves
from gemseo.post.dataset.parallel_coordinates import ParallelCoordinates
from gemseo.post.dataset.radviz import Radar
from gemseo.post.dataset.scatter_plot_matrix import ScatterMatrix
from matplotlib import pyplot as plt
from numpy.random import choice
configure_logger()
<RootLogger root (INFO)>
Load Iris dataset¶
We can easily load this dataset by means of the
load_dataset() function of the API:
iris = load_dataset("IrisDataset")
and get some information about it
print(iris)
Iris
Number of samples: 150
Number of variables: 5
Variables names and sizes by group:
labels: specy (1)
parameters: petal_length (1), petal_width (1), sepal_length (1), sepal_width (1)
Number of dimensions (total = 5) by group:
labels: 1
parameters: 4
Manipulate the dataset¶
We randomly select 10 samples to display.
shown_samples = choice(iris.length, size=10, replace=False)
If the pandas library is installed, we can export the iris dataset to a dataframe and print(it.
dataframe = iris.export_to_dataframe()
print(dataframe)
parameters labels
petal_length petal_width sepal_length sepal_width specy
0 0 0 0 0
0 1.4 0.2 5.1 3.5 0.0
1 1.4 0.2 4.9 3.0 0.0
2 1.3 0.2 4.7 3.2 0.0
3 1.5 0.2 4.6 3.1 0.0
4 1.4 0.2 5.0 3.6 0.0
.. ... ... ... ... ...
145 5.2 2.3 6.7 3.0 2.0
146 5.0 1.9 6.3 2.5 2.0
147 5.2 2.0 6.5 3.0 2.0
148 5.4 2.3 6.2 3.4 2.0
149 5.1 1.8 5.9 3.0 2.0
[150 rows x 5 columns]
We can also easily access the 10 samples previously selected, either globally
data = iris.get_all_data(False)
print(data[0][shown_samples, :])
[[0. 5. 3.2 1.2 0.2]
[2. 6.8 3.2 5.9 2.3]
[1. 6. 2.2 4. 1. ]
[2. 6. 3. 4.8 1.8]
[1. 6. 3.4 4.5 1.6]
[0. 4.9 3.1 1.5 0.2]
[2. 7.3 2.9 6.3 1.8]
[2. 6.2 3.4 5.4 2.3]
[0. 5.4 3.4 1.7 0.2]
[2. 5.8 2.7 5.1 1.9]]
or only the parameters:
parameters = iris.get_data_by_group("parameters")
print(parameters[shown_samples, :])
[[5. 3.2 1.2 0.2]
[6.8 3.2 5.9 2.3]
[6. 2.2 4. 1. ]
[6. 3. 4.8 1.8]
[6. 3.4 4.5 1.6]
[4.9 3.1 1.5 0.2]
[7.3 2.9 6.3 1.8]
[6.2 3.4 5.4 2.3]
[5.4 3.4 1.7 0.2]
[5.8 2.7 5.1 1.9]]
or only the labels:
labels = iris.get_data_by_group("labels")
print(labels[shown_samples, :])
[[0.]
[2.]
[1.]
[2.]
[1.]
[0.]
[2.]
[2.]
[0.]
[2.]]
Plot the dataset¶
Lastly, we can plot the dataset in various ways. We will note that the samples are colored according to their labels.
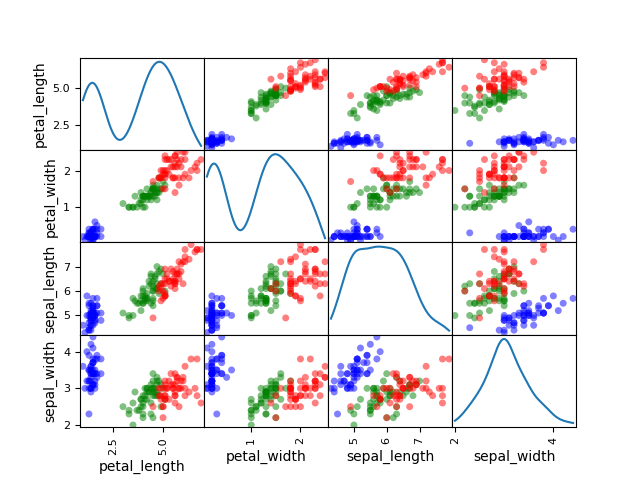
Plot scatter matrix¶
We can use the ScatterMatrix plot where each non-diagonal block
represents the samples according to the x- and y- coordinates names
while the diagonal ones approximate the probability distributions of the
variables, using either an histogram or a kernel-density estimator.
ScatterMatrix(iris, classifier="specy", kde=True).execute(save=False, show=False)
/home/docs/checkouts/readthedocs.org/user_builds/gemseo/envs/4.1.0/lib/python3.9/site-packages/gemseo/post/dataset/scatter_plot_matrix.py:135: UserWarning: To output multiple subplots, the figure containing the passed axes is being cleared.
sub_axes = scatter_matrix(
[<Figure size 640x480 with 16 Axes>]
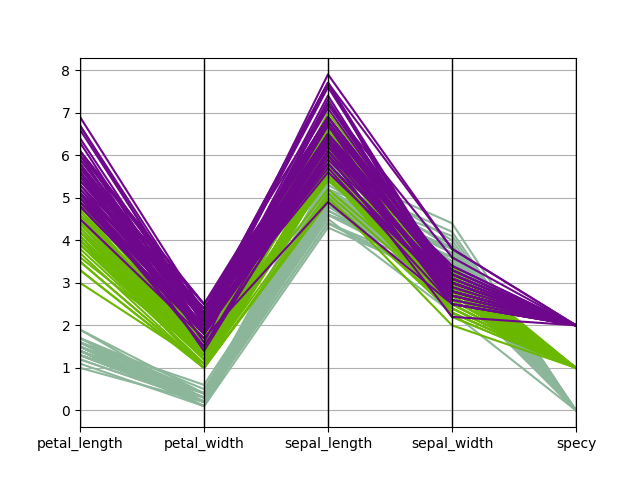
Plot parallel coordinates¶
We can use the
ParallelCoordinates plot,
a.k.a. cowebplot, where each samples
is represented by a continuous straight line in pieces whose nodes are
indexed by the variables names and measure the variables values.
ParallelCoordinates(iris, "specy").execute(save=False, show=False)
[<Figure size 640x480 with 1 Axes>]
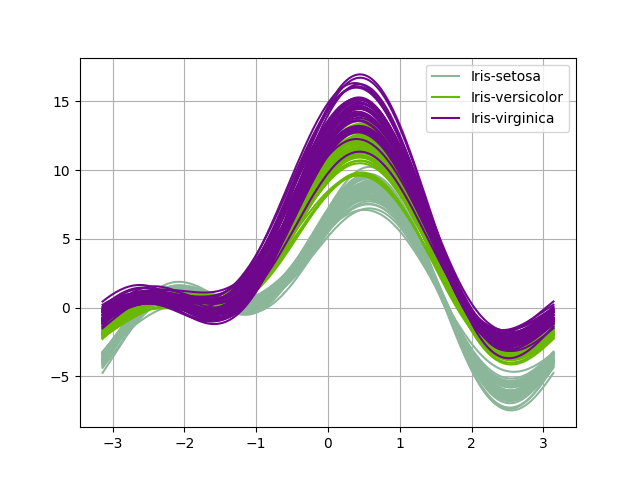
Plot Andrews curves¶
We can use the AndrewsCurves plot
which can be viewed as a smooth
version of the parallel coordinates. Each sample is represented by a curve
and if there is structure in data, it may be visible in the plot.
AndrewsCurves(iris, "specy").execute(save=False, show=False)
[<Figure size 640x480 with 1 Axes>]
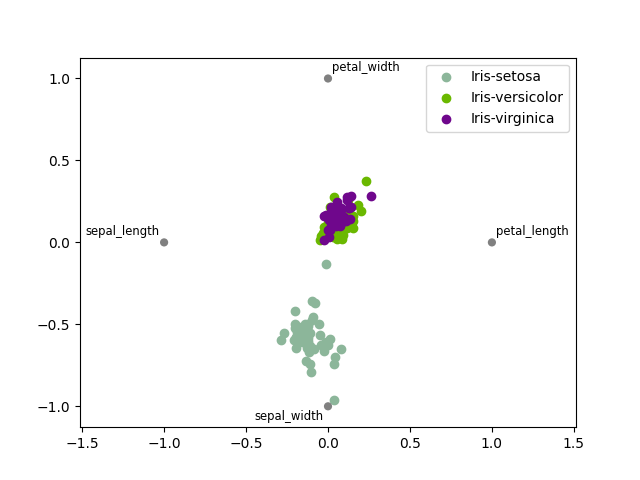
Plot Radar¶
We can use the Radar plot
Radar(iris, "specy").execute(save=False, show=False)
# Workaround for HTML rendering, instead of ``show=True``
plt.show()
Total running time of the script: ( 0 minutes 1.546 seconds)
