Note
Click here to download the full example code
Sobol’ analysis¶
from __future__ import annotations
import pprint
from gemseo.algos.parameter_space import ParameterSpace
from gemseo.api import create_discipline
from gemseo.uncertainty.sensitivity.sobol.analysis import SobolAnalysis
from matplotlib import pyplot as plt
from numpy import pi
In this example, we consider a function from \([-\pi,\pi]^3\) to \(\mathbb{R}^3\):
where \(f(a,b,c)=\sin(a)+7\sin(b)^2+0.1*c^4\sin(a)\) is the Ishigami function:
expressions = {
"y1": "sin(x1)+7*sin(x2)**2+0.1*x3**4*sin(x1)",
"y2": "sin(x2)+7*sin(x1)**2+0.1*x3**4*sin(x2)",
}
discipline = create_discipline(
"AnalyticDiscipline", expressions=expressions, name="Ishigami2"
)
Then, we consider the case where the deterministic variables \(x_1\), \(x_2\) and \(x_3\) are replaced by the uncertain variables \(X_1\), \(X_2\) and \(X_3\). The latter are independent and identically distributed according to the uniform distribution between \(-\pi\) and \(\pi\):
uncertain_space = ParameterSpace()
for variable_name in ["x1", "x2", "x3"]:
uncertain_space.add_random_variable(
variable_name, "OTUniformDistribution", minimum=-pi, maximum=pi
)
From that,
we would like to carry out a sensitivity analysis with the random outputs
\(Y_1=f(X_1,X_2,X_3)\) and \(Y_2=f(X_2,X_1,X_3)\).
For that,
we can compute the sensitivity indices from a SobolAnalysis:
sobol_analysis = SobolAnalysis([discipline], uncertain_space, 100)
sobol_analysis.main_method = "total"
sobol_analysis.compute_indices()
{'first': {'y2': [{'x1': array([0.75387699]), 'x2': array([0.26516667]), 'x3': array([0.35758815])}], 'y1': [{'x1': array([0.19426511]), 'x2': array([0.21211854]), 'x3': array([0.01782884])}]}, 'total': {'y2': [{'x1': array([0.36991381]), 'x2': array([0.60974968]), 'x3': array([0.34776012])}], 'y1': [{'x1': array([0.87052721]), 'x2': array([0.37726189]), 'x3': array([0.29945874])}]}}
The resulting indices are the first-order and total Sobol’ indices:
pprint.pprint(sobol_analysis.indices)
{'first': {'y1': [{'x1': array([0.19426511]),
'x2': array([0.21211854]),
'x3': array([0.01782884])}],
'y2': [{'x1': array([0.75387699]),
'x2': array([0.26516667]),
'x3': array([0.35758815])}]},
'total': {'y1': [{'x1': array([0.87052721]),
'x2': array([0.37726189]),
'x3': array([0.29945874])}],
'y2': [{'x1': array([0.36991381]),
'x2': array([0.60974968]),
'x3': array([0.34776012])}]}}
They can also be accessed separately:
pprint.pprint(sobol_analysis.first_order_indices)
pprint.pprint(sobol_analysis.total_order_indices)
{'y1': [{'x1': array([0.19426511]),
'x2': array([0.21211854]),
'x3': array([0.01782884])}],
'y2': [{'x1': array([0.75387699]),
'x2': array([0.26516667]),
'x3': array([0.35758815])}]}
{'y1': [{'x1': array([0.87052721]),
'x2': array([0.37726189]),
'x3': array([0.29945874])}],
'y2': [{'x1': array([0.36991381]),
'x2': array([0.60974968]),
'x3': array([0.34776012])}]}
One can also obtain their confidence intervals:
pprint.pprint(sobol_analysis.get_intervals())
pprint.pprint(sobol_analysis.get_intervals(first_order=False))
{'y1': [{'x1': array([-0.03400625, 0.54199216]),
'x2': array([-0.00512583, 0.66154825]),
'x3': array([-0.40647314, 0.44110609])}],
'y2': [{'x1': array([0.10944697, 1.99005049]),
'x2': array([-0.31379805, 0.99720365]),
'x3': array([-0.43222646, 1.8321417 ])}]}
{'y1': [{'x1': array([0.50048303, 1.91420643]),
'x2': array([-0.35083774, 1.67786025]),
'x3': array([-0.19380841, 1.59540502])}],
'y2': [{'x1': array([-0.63775393, 3.12446122]),
'x2': array([-0.01837286, 2.23112503]),
'x3': array([-0.21486175, 1.97989239])}]}
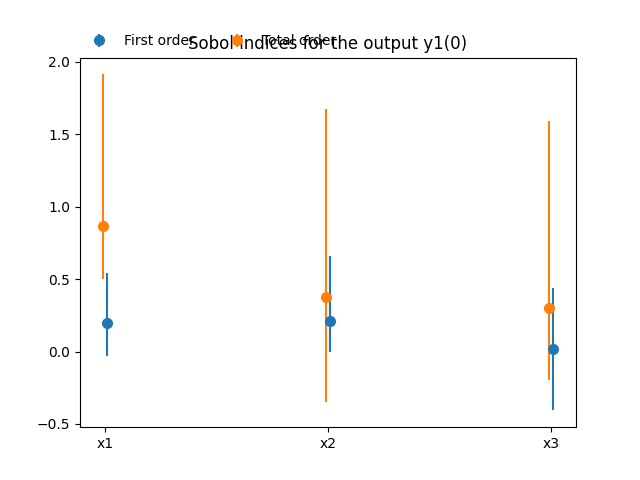
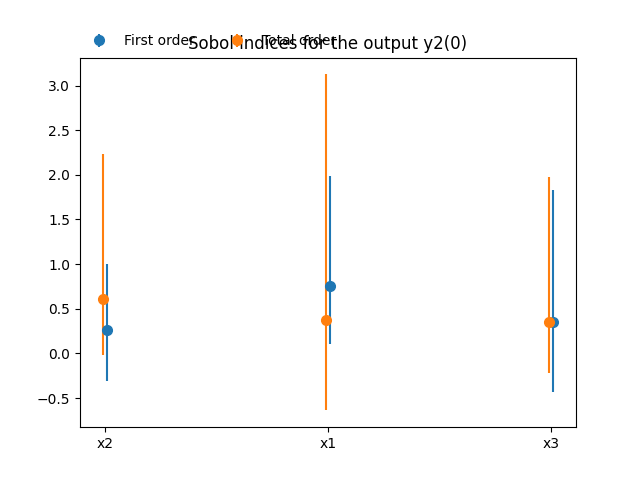
The main indices are the total Sobol’ indices
(SobolAnalysis.main_method can also be set to “first”
to use the first-order indices as main indices):
pprint.pprint(sobol_analysis.main_indices)
{'y1': [{'x1': array([0.87052721]),
'x2': array([0.37726189]),
'x3': array([0.29945874])}],
'y2': [{'x1': array([0.36991381]),
'x2': array([0.60974968]),
'x3': array([0.34776012])}]}
These main indices are used to sort the input parameters by decreasing order of influence. We can observe that this ranking is not the same for both outputs:
print(sobol_analysis.sort_parameters("y1"))
print(sobol_analysis.sort_parameters("y2"))
['x1', 'x2', 'x3']
['x2', 'x1', 'x3']
Lastly,
we can use the method SobolAnalysis.plot()
to visualize both first-order and total Sobol’ indices:
sobol_analysis.plot("y1", save=False, show=False)
sobol_analysis.plot("y2", save=False, show=False)
# Workaround for HTML rendering, instead of ``show=True``
plt.show()
Total running time of the script: ( 0 minutes 0.458 seconds)