api module¶
Introduction¶
Here is the Application Programming Interface (API) of GEMSEO, a set of high level functions for ease of use.
Make GEMSEO ever more accessible¶
The aim of this API is to provide high level functions that are sufficient to use GEMSEO in most cases, without requiring a deep knowledge of GEMSEO.
Besides, these functions shall change much less often than the internal classes, which is key for backward compatibility, which means ensuring that your current scripts using GEMSEO will be usable with the future versions of GEMSEO.
Connect GEMSEO to your favorite tools¶
The API also facilitates the interfacing of GEMSEO with a platform or other software.
To interface a simulation software with GEMSEO, please refer to: Interfacing simulation software.
Extending GEMSEO¶
Table of contents¶
Algorithms¶
Cache¶
Configuration¶
Coupling¶
Design space¶
Disciplines¶
Formulations¶
MDA¶
Post-processing¶
Scalable¶
Scenario¶
Surrogates¶
API functions¶
- gemseo.api.AlgorithmFeatures¶
alias of
AlgorithmFeature
- gemseo.api.compute_doe(variables_space, algo_name, size=None, unit_sampling=False, **options)[source]¶
Compute a design of experiments (DOE) in a variables space.
- Parameters:
variables_space (DesignSpace) – The variables space to be sampled.
size (int | None) – The size of the DOE. If
None, the size is deduced from theoptions.algo_name (str) – The DOE algorithm.
unit_sampling (bool) –
Whether to sample in the unit hypercube.
By default it is set to False.
**options (DOELibraryOptionType) – The options of the DOE algorithm.
- Returns:
The design of experiments whose rows are the samples and columns the variables.
- Return type:
ndarray
Examples
>>> from gemseo.api import compute_doe, create_design_space >>> variables_space = create_design_space() >>> variables_space.add_variable("x", 2, l_b=-1.0, u_b=1.0) >>> doe = compute_doe(variables_space, algo_name="lhs", size=5)
- gemseo.api.configure(activate_discipline_counters=True, activate_function_counters=True, activate_progress_bar=True, activate_discipline_cache=True, check_input_data=True, check_output_data=True, check_desvars_bounds=True)[source]¶
Update the configuration of GEMSEO if needed.
This could be useful to speed up calculations in presence of cheap disciplines such as analytic formula and surrogate models.
Warning
This function should be called before calling anything from GEMSEO.
- Parameters:
activate_discipline_counters (bool) –
Whether to activate the counters attached to the disciplines, in charge of counting their execution time, number of evaluations and number of linearizations.
By default it is set to True.
activate_function_counters (bool) –
Whether to activate the counters attached to the functions, in charge of counting their number of evaluations.
By default it is set to True.
activate_progress_bar (bool) –
Whether to activate the progress bar attached to the drivers, in charge to log the execution of the process: iteration, execution time and objective value.
By default it is set to True.
activate_discipline_cache (bool) –
Whether to activate the discipline cache.
By default it is set to True.
check_input_data (bool) –
Whether to check the input data of a discipline before execution.
By default it is set to True.
check_output_data (bool) –
Whether to check the output data of a discipline before execution.
By default it is set to True.
check_desvars_bounds (bool) –
Whether to check the membership of design variables in the bounds when evaluating the functions in OptimizationProblem.
By default it is set to True.
- Return type:
None
- gemseo.api.configure_logger(logger_name=None, level=20, date_format='%H:%M:%S', message_format='%(levelname)8s - %(asctime)s: %(message)s', filename=None, filemode='a')[source]¶
Configure GEMSEO logging.
- Parameters:
logger_name (str | None) – The name of the logger to configure. If
None, return the root logger.The numerical value or name of the logging level, as defined in
logging. Values can either belogging.NOTSET("NOTSET"),logging.DEBUG("DEBUG"),logging.INFO("INFO"),logging.WARNING("WARNING"or"WARN"),logging.ERROR("ERROR"), orlogging.CRITICAL("FATAL"or"CRITICAL").By default it is set to 20.
date_format (str) –
The logging date format.
By default it is set to “%H:%M:%S”.
message_format (str) –
The logging message format.
By default it is set to “%(levelname)8s - %(asctime)s: %(message)s”.
filename (str | Path | None) – The path to the log file, if outputs must be written in a file.
filemode (str) –
The logging output file mode, either ‘w’ (overwrite) or ‘a’ (append).
By default it is set to “a”.
- Return type:
Logger
Examples
>>> import logging >>> configure_logger(level=logging.WARNING)
- gemseo.api.create_cache(cache_type, name=None, **options)[source]¶
Return a cache.
- Parameters:
- Returns:
The cache.
- Return type:
Examples
>>> from gemseo.api import create_cache >>> cache = create_cache('MemoryFullCache') >>> print(cache) +--------------------------------+ | MemoryFullCache | +--------------+-----------------+ | Property | Value | +--------------+-----------------+ | Type | MemoryFullCache | | Tolerance | 0.0 | | Input names | None | | Output names | None | | Length | 0 | +--------------+-----------------+
- gemseo.api.create_dataset(name, data, variables=None, sizes=None, groups=None, by_group=True, delimiter=',', header=True, default_name=None)[source]¶
Create a dataset from a NumPy array or a data file.
- Parameters:
name (str) – The name to be given to the dataset.
data (ndarray | str | Path) – The data to be stored in the dataset, either a NumPy array or a file path.
variables (list[str] | None) – The names of the variables. If
Noneand header is True, read the names from the first line of the file. IfNoneand header is False, use default names based on the patterns theDataset.DEFAULT_NAMESassociated with the different groups.sizes (dict[str, int] | None) – The sizes of the variables. If
None, assume that all the variables have a size equal to 1.groups (dict[str, str] | None) – The groups of the variables. If
None, useDataset.DEFAULT_GROUPfor all the variables.by_group (bool) –
If True, store the data by group. Otherwise, store them by variables.
By default it is set to True.
delimiter (str) –
The field delimiter.
By default it is set to “,”.
header (bool) –
If True and data is a string, read the variables names on the first line of the file.
By default it is set to True.
default_name (str | None) – The name of the variable to be used as a pattern when variables is None. If
None, use theDataset.DEFAULT_NAMESfor this group if it exists. Otherwise, use the group name.
- Returns:
The dataset generated from the NumPy array or data file.
- Return type:
See also
- gemseo.api.create_design_space()[source]¶
Create an empty design space.
- Returns:
An empty design space.
- Return type:
Examples
>>> from gemseo.api import create_design_space >>> design_space = create_design_space() >>> design_space.add_variable('x', l_b=-1, u_b=1, value=0.) >>> print(design_space) Design Space: +------+-------------+-------+-------------+-------+ | name | lower_bound | value | upper_bound | type | +------+-------------+-------+-------------+-------+ | x | -1 | 0 | 1 | float | +------+-------------+-------+-------------+-------+
- gemseo.api.create_discipline(discipline_name, **options)[source]¶
Instantiate one or more disciplines.
- Parameters:
- Returns:
The disciplines.
Examples
>>> from gemseo.api import create_discipline >>> discipline = create_discipline('Sellar1') >>> discipline.execute() {'x_local': array([0.+0.j]), 'x_shared': array([1.+0.j, 0.+0.j]), 'y_0': array([0.89442719+0.j]), 'y_1': array([1.+0.j])}
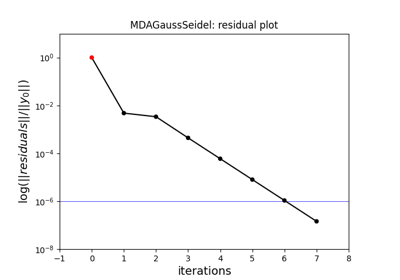
- gemseo.api.create_mda(mda_name, disciplines, **options)[source]¶
Create a multidisciplinary analysis (MDA).
- Parameters:
mda_name (str) – The name of the MDA.
disciplines (Sequence[MDODiscipline]) – The disciplines.
**options (Any) – The options of the MDA.
- Returns:
The MDA.
- Return type:
Examples
>>> from gemseo.api import create_discipline, create_mda >>> disciplines = create_discipline(['Sellar1', 'Sellar2']) >>> mda = create_mda('MDAGaussSeidel', disciplines) >>> mda.execute() {'x_local': array([0.+0.j]), 'x_shared': array([1.+0.j, 0.+0.j]), 'y_0': array([0.79999995+0.j]), 'y_1': array([1.79999995+0.j])}
See also
- gemseo.api.create_parameter_space()[source]¶
Create an empty parameter space.
- Returns:
An empty parameter space.
- Return type:
- gemseo.api.create_scalable(name, data, sizes=None, **parameters)[source]¶
Create a scalable discipline from a dataset.
- Parameters:
- Returns:
The scalable discipline.
- Return type:
- gemseo.api.create_scenario(disciplines, formulation, objective_name, design_space, name=None, scenario_type='MDO', grammar_type='JSONGrammar', maximize_objective=False, **options)[source]¶
Initialize a scenario.
- Parameters:
disciplines (Sequence[MDODiscipline]) – The disciplines used to compute the objective, constraints and observables from the design variables.
formulation (str) – The class name of the
MDOFormulation, e.g."MDF","IDF"or"BiLevel".objective_name (str) – The name(s) of the discipline output(s) used as objective. If multiple names are passed, the objective will be a vector.
design_space (DesignSpace | str | Path) – The search space including at least the design variables (some formulations requires additional variables, e.g.
IDFwith the coupling variables).name (str | None) – The name to be given to this scenario. If
None, use the name of the class.scenario_type (str) –
The type of the scenario, e.g.
"MDO"or"DOE".By default it is set to “MDO”.
grammar_type (str) –
The type of grammar to declare the input and output variables either
JSON_GRAMMAR_TYPEorSIMPLE_GRAMMAR_TYPE.By default it is set to “JSONGrammar”.
maximize_objective (bool) –
Whether to maximize the objective.
By default it is set to False.
**options (Any) – The options of the
MDOFormulation.
- Return type:
Examples
>>> from gemseo.api import create_discipline, create_scenario >>> from gemseo.problems.sellar.sellar_design_space import SellarDesignSpace >>> disciplines = create_discipline(['Sellar1', 'Sellar2', 'SellarSystem']) >>> design_space = SellarDesignSpace() >>> scenario = create_scenario(disciplines, 'MDF', 'obj', design_space, 'SellarMDFScenario')
- gemseo.api.create_surrogate(surrogate, data=None, transformer=mappingproxy({'inputs': <gemseo.mlearning.transform.scaler.min_max_scaler.MinMaxScaler object>, 'outputs': <gemseo.mlearning.transform.scaler.min_max_scaler.MinMaxScaler object>}), disc_name=None, default_inputs=None, input_names=None, output_names=None, **parameters)[source]¶
Create a surrogate discipline, either from a dataset or a regression model.
- Parameters:
surrogate (str | MLRegressionAlgo) – Either the class name or the instance of the
MLRegressionAlgo.data (Dataset | None) – The learning dataset to train the regression model. If
None, the regression model is supposed to be trained.transformer (TransformerType) –
The strategies to transform the variables. The values are instances of
Transformerwhile the keys are the names of either the variables or the groups of variables, e.g. “inputs” or “outputs” in the case of the regression algorithms. If a group is specified, theTransformerwill be applied to all the variables of this group. If None, do not transform the variables. TheMLRegressionAlgo.DEFAULT_TRANSFORMERuses theMinMaxScalerstrategy for both input and output variables.By default it is set to {‘inputs’: <gemseo.mlearning.transform.scaler.min_max_scaler.MinMaxScaler object at 0x7fbc6ba6e520>, ‘outputs’: <gemseo.mlearning.transform.scaler.min_max_scaler.MinMaxScaler object at 0x7fbc6ba6e4c0>}.
disc_name (str | None) – The name to be given to the surrogate discipline. If
None, concatenateSHORT_ALGO_NAMEanddata.name.default_inputs (dict[str, ndarray] | None) – The default values of the inputs. If
None, use the center of the learning input space.input_names (Iterable[str] | None) – The names of the input variables. If
None, consider all input variables mentioned in the learning dataset.output_names (Iterable[str] | None) – The names of the output variables. If
None, consider all input variables mentioned in the learning dataset.**parameters (Any) – The parameters of the machine learning algorithm.
- Return type:
- gemseo.api.execute_algo(opt_problem, algo_name, algo_type='opt', **options)[source]¶
Solve an optimization problem.
- Parameters:
opt_problem (OptimizationProblem) – The optimization problem to be solved.
algo_name (str) – The name of the algorithm to be used to solve optimization problem.
algo_type (str) –
The type of algorithm, either “opt” for optimization or “doe” for design of experiments.
By default it is set to “opt”.
**options (Any) – The options of the algorithm.
- Return type:
Examples
>>> from gemseo.api import execute_algo >>> from gemseo.problems.analytical.rosenbrock import Rosenbrock >>> opt_problem = Rosenbrock() >>> opt_result = execute_algo(opt_problem, 'SLSQP') >>> opt_result Optimization result: |_ Design variables: [0.99999787 0.99999581] |_ Objective function: 5.054173713127532e-12 |_ Feasible solution: True
- gemseo.api.execute_post(to_post_proc, post_name, **options)[source]¶
Post-process a result.
- Parameters:
to_post_proc (Scenario | OptimizationProblem | str | Path) – The result to be post-processed, either a DOE scenario, a MDO scenario, an optimization problem or a path to an HDF file containing a saved optimization problem.
post_name (str) – The name of the post-processing.
**options (Any) – The post-processing options.
- Returns:
The figures, to be customized if not closed.
- Return type:
Examples
>>> from gemseo.api import create_discipline, create_scenario, execute_post >>> from gemseo.problems.sellar.sellar_design_space import SellarDesignSpace >>> disciplines = create_discipline(['Sellar1', 'Sellar2', 'SellarSystem']) >>> design_space = SellarDesignSpace() >>> scenario = create_scenario(disciplines, 'MDF', 'obj', design_space, 'SellarMDFScenario') >>> scenario.execute({'algo': 'NLOPT_SLSQP', 'max_iter': 100}) >>> execute_post(scenario, 'OptHistoryView', show=False, save=True)
- gemseo.api.export_design_space(design_space, output_file, export_hdf=False, fields=None, header_char='', **table_options)[source]¶
Save a design space to a text or HDF file.
- Parameters:
design_space (DesignSpace) – The design space to be saved.
output_file (str | Path) – The path to the file.
export_hdf (bool) –
If True, save to an HDF file. Otherwise, save to a text file.
By default it is set to False.
fields (Sequence[str] | None) – The fields to be exported. If
None, export all fields.header_char (str) –
The header character.
By default it is set to “”.
**table_options (Any) – The names and values of additional attributes for the
PrettyTableview generated byDesignSpace.get_pretty_table().
- Return type:
None
Examples
>>> from gemseo.api import create_design_space, export_design_space >>> design_space = create_design_space() >>> design_space.add_variable('x', l_b=-1, u_b=1, value=0.) >>> export_design_space(design_space, 'file.txt')
See also
- gemseo.api.generate_coupling_graph(disciplines, file_path='coupling_graph.pdf', full=True)[source]¶
Generate a graph of the couplings between disciplines.
- Parameters:
disciplines (Sequence[MDODiscipline]) – The disciplines from which the graph is generated.
file_path (str | Path) –
The path of the file to save the figure.
By default it is set to “coupling_graph.pdf”.
full (bool) –
If True, generate the full coupling graph. Otherwise, generate the condensed one.
By default it is set to True.
- Return type:
None
Examples
>>> from gemseo.api import create_discipline, generate_coupling_graph >>> disciplines = create_discipline(['Sellar1', 'Sellar2', 'SellarSystem']) >>> generate_coupling_graph(disciplines)
See also
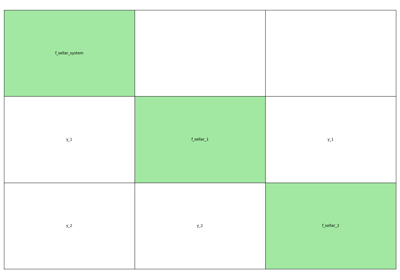
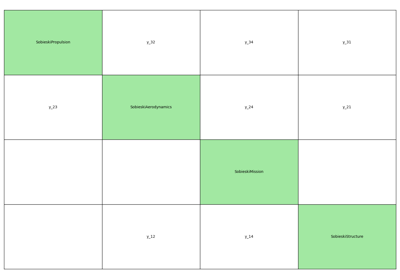
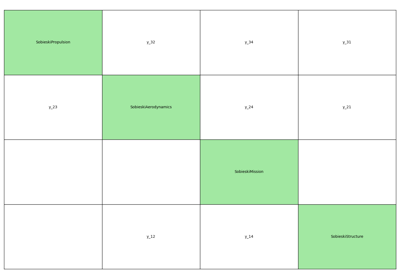
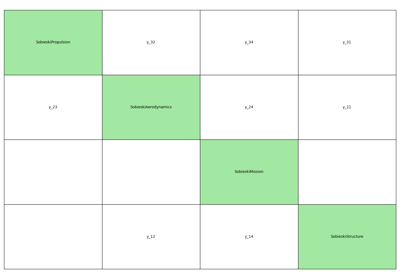
- gemseo.api.generate_n2_plot(disciplines, file_path='n2.pdf', show_data_names=True, save=True, show=False, fig_size=(15.0, 10.0), open_browser=False)[source]¶
Generate a N2 plot from disciplines.
It can be static (e.g. PDF, PNG, …), dynamic (HTML) or both.
The disciplines are located on the diagonal of the N2 plot while the coupling variables are situated on the other blocks of the matrix view. A coupling variable is outputted by a discipline horizontally and enters another vertically. On the static plot, a blue diagonal block represent a self-coupled discipline, i.e. a discipline having some of its outputs as inputs.
- Parameters:
disciplines (Sequence[MDODiscipline]) – The disciplines from which the N2 chart is generated.
file_path (str | Path) –
The file path to save the static N2 chart. show_data_names: Whether to show the names of the coupling variables
between two disciplines; otherwise, circles are drawn, whose size depends on the number of coupling names.
By default it is set to “n2.pdf”.
save (bool) –
Whether to save the static N2 chart.
By default it is set to True.
show (bool) –
Whether to show the static N2 chart.
By default it is set to False.
fig_size (tuple[float, float]) –
The width and height of the static N2 chart.
By default it is set to (15.0, 10.0).
open_browser (bool) –
Whether to display the interactive N2 chart in a browser.
By default it is set to False.
show_data_names (bool) –
By default it is set to True.
- Return type:
None
Examples
>>> from gemseo.api import create_discipline, generate_n2_plot >>> disciplines = create_discipline(['Sellar1', 'Sellar2', 'SellarSystem']) >>> generate_n2_plot(disciplines)
See also
- gemseo.api.get_algorithm_features(algorithm_name)[source]¶
Return the features of an optimization algorithm.
- Parameters:
algorithm_name (str) – The name of the optimization algorithm.
- Returns:
The features of the optimization algorithm.
- Raises:
ValueError – When the optimization algorithm does not exist.
- Return type:
AlgorithmFeature
- gemseo.api.get_algorithm_options_schema(algorithm_name, output_json=False, pretty_print=False)[source]¶
Return the schema of the options of an algorithm.
- Parameters:
- Returns:
The schema of the options of the algorithm.
- Raises:
ValueError – When the algorithm is not available.
- Return type:
Examples
>>> from gemseo.api import get_algorithm_options_schema >>> schema = get_algorithm_options_schema('NLOPT_SLSQP', pretty_print=True)
- gemseo.api.get_available_caches()[source]¶
Return the names of the available caches.
Examples
>>> from gemseo.api import get_available_caches >>> get_available_caches() ['AbstractFullCache', 'HDF5Cache', 'MemoryFullCache', 'SimpleCache']
- gemseo.api.get_available_disciplines()[source]¶
Return the names of the available disciplines.
Examples
>>> from gemseo.api import get_available_disciplines >>> print(get_available_disciplines()) ['RosenMF', 'SobieskiAerodynamics', 'ScalableKriging', 'DOEScenario', 'MDOScenario', 'SobieskiMission', 'SobieskiDiscipline', 'Sellar1', 'Sellar2', 'MDOChain', 'SobieskiStructure', 'AutoPyDiscipline', 'Structure', 'SobieskiPropulsion', 'Scenario', 'AnalyticDiscipline', 'MDOScenarioAdapter', 'ScalableDiscipline', 'SellarSystem', 'Aerodynamics', 'Mission', 'PropaneComb1', 'PropaneComb2', 'PropaneComb3', 'PropaneReaction', 'MDOParallelChain']
- gemseo.api.get_available_doe_algorithms()[source]¶
Return the names of the available design of experiments (DOEs) algorithms.
Examples
>>> from gemseo.api import get_available_doe_algorithms >>> get_available_doe_algorithms()
- gemseo.api.get_available_formulations()[source]¶
Return the names of the available formulations.
Examples
>>> from gemseo.api import get_available_formulations >>> get_available_formulations()
- gemseo.api.get_available_mdas()[source]¶
Return the names of the available multidisciplinary analyses (MDAs).
Examples
>>> from gemseo.api import get_available_mdas >>> get_available_mdas()
See also
- gemseo.api.get_available_opt_algorithms()[source]¶
Return the names of the available optimization algorithms.
Examples
>>> from gemseo.api import get_available_opt_algorithms >>> get_available_opt_algorithms()
- gemseo.api.get_available_post_processings()[source]¶
Return the names of the available optimization post-processings.
Examples
>>> from gemseo.api import get_available_post_processings >>> print(get_available_post_processings()) ['ScatterPlotMatrix', 'VariableInfluence', 'ConstraintsHistory', 'RadarChart', 'Robustness', 'Correlations', 'SOM', 'KMeans', 'ParallelCoordinates', 'GradientSensitivity', 'OptHistoryView', 'BasicHistory', 'ObjConstrHist', 'QuadApprox']
- gemseo.api.get_available_scenario_types()[source]¶
Return the names of the available scenario types.
Examples
>>> from gemseo.api import get_available_scenario_types >>> get_available_scenario_types()
- gemseo.api.get_available_surrogates()[source]¶
Return the names of the available surrogate disciplines.
Examples
>>> from gemseo.api import get_available_surrogates >>> print(get_available_surrogates()) ['RBFRegressor', 'GaussianProcessRegressor', 'LinearRegressor', 'PCERegressor']
See also
- gemseo.api.get_discipline_inputs_schema(discipline, output_json=False, pretty_print=False)[source]¶
Return the schema of the inputs of a discipline.
- Parameters:
discipline (MDODiscipline) – The discipline.
output_json (bool) –
Whether to apply the JSON format to the schema.
By default it is set to False.
pretty_print (bool) –
Whether to print the schema in a tabular way.
By default it is set to False.
- Returns:
The schema of the inputs of the discipline.
- Return type:
Examples
>>> from gemseo.api import create_discipline, get_discipline_inputs_schema >>> discipline = create_discipline('Sellar1') >>> schema = get_discipline_inputs_schema(discipline, pretty_print=True)
- gemseo.api.get_discipline_options_defaults(discipline_name)[source]¶
Return the default values of the options of a discipline.
- Parameters:
discipline_name (str) – The name of the discipline.
- Returns:
The default values of the options of the discipline.
- Return type:
Examples
>>> from gemseo.api import get_discipline_options_defaults >>> get_discipline_options_defaults('Sellar1')
- gemseo.api.get_discipline_options_schema(discipline_name, output_json=False, pretty_print=False)[source]¶
Return the schema of a discipline.
- Parameters:
- Returns:
The schema of the discipline.
- Return type:
Examples
>>> from gemseo.api import get_discipline_options_schema >>> schema = get_discipline_options_schema('Sellar1', pretty_print=True)
- gemseo.api.get_discipline_outputs_schema(discipline, output_json=False, pretty_print=False)[source]¶
Return the schema of the outputs of a discipline.
- Parameters:
discipline (MDODiscipline) – The discipline.
output_json (bool) –
Whether to apply the JSON format to the schema.
By default it is set to False.
pretty_print (bool) –
Whether to print the schema in a tabular way.
By default it is set to False.
- Returns:
The schema of the outputs of the discipline.
- Return type:
Examples
>>> from gemseo.api import get_discipline_outputs_schema, create_discipline >>> discipline = create_discipline('Sellar1') >>> get_discipline_outputs_schema(discipline, pretty_print=True)
- gemseo.api.get_formulation_options_schema(formulation_name, output_json=False, pretty_print=False)[source]¶
Return the schema of the options of a formulation.
- Parameters:
- Returns:
The schema of the options of the formulation.
- Return type:
Examples
>>> from gemseo.api import get_formulation_options_schema >>> schema = get_formulation_options_schema('MDF', pretty_print=True)
- gemseo.api.get_formulation_sub_options_schema(formulation_name, output_json=False, pretty_print=False, **formulation_options)[source]¶
Return the schema of the sub-options of a formulation.
- Parameters:
formulation_name (str) – The name of the formulation.
output_json (bool) –
Whether to apply the JSON format to the schema.
By default it is set to False.
pretty_print (bool) –
Whether to print the schema in a tabular way.
By default it is set to False.
**formulation_options (Any) – The options of the formulation required for its instantiation.
- Returns:
The schema of the sub-options of the formulation.
- Return type:
Examples
>>> from gemseo.api import get_formulation_sub_options_schema >>> schema = get_formulation_sub_options_schema('MDF', >>> main_mda_name='MDAJacobi', >>> pretty_print=True)
- gemseo.api.get_formulations_options_defaults(formulation_name)[source]¶
Return the default values of the options of a formulation.
- Parameters:
formulation_name (str) – The name of the formulation.
- Returns:
The default values of the options of the formulation.
- Return type:
Examples
>>> from gemseo.api import get_formulations_options_defaults >>> get_formulations_options_defaults('MDF') {'main_mda_name': 'MDAChain', 'maximize_objective': False, 'inner_mda_name': 'MDAJacobi'}
- gemseo.api.get_formulations_sub_options_defaults(formulation_name, **formulation_options)[source]¶
Return the default values of the sub-options of a formulation.
- Parameters:
- Returns:
The default values of the sub-options of the formulation.
- Return type:
Examples
>>> from gemseo.api import get_formulations_sub_options_defaults >>> get_formulations_sub_options_defaults('MDF', >>> main_mda_name='MDAJacobi')
- gemseo.api.get_mda_options_schema(mda_name, output_json=False, pretty_print=False)[source]¶
Return the schema of the options of a multidisciplinary analysis (MDA).
- Parameters:
- Returns:
The schema of the options of the MDA.
- Return type:
Examples
>>> from gemseo.api import get_mda_options_schema >>> get_mda_options_schema('MDAJacobi')
See also
- gemseo.api.get_post_processing_options_schema(post_proc_name, output_json=False, pretty_print=False)[source]¶
Return the schema of the options of a post-processing.
- Parameters:
- Returns:
The schema of the options of the post-processing.
- Return type:
Examples
>>> from gemseo.api import get_post_processing_options_schema >>> schema = get_post_processing_options_schema('OptHistoryView', >>> pretty_print=True)
See also
- gemseo.api.get_scenario_differentiation_modes()[source]¶
Return the names of the available differentiation modes of a scenario.
- Returns:
The names of the available differentiation modes of a scenario.
Examples
>>> from gemseo.api import get_scenario_differentiation_modes >>> get_scenario_differentiation_modes()
- gemseo.api.get_scenario_inputs_schema(scenario, output_json=False, pretty_print=False)[source]¶
Return the schema of the inputs of a scenario.
- Parameters:
- Returns:
The schema of the inputs of the scenario.
- Return type:
Examples
>>> from gemseo.api import create_discipline, create_scenario, get_scenario_inputs_schema >>> from gemseo.problems.sellar.sellar_design_space import SellarDesignSpace >>> design_space = SellarDesignSpace() >>> disciplines = create_discipline(['Sellar1','Sellar2','SellarSystem']) >>> scenario = create_scenario(disciplines, 'MDF', 'obj', design_space, 'my_scenario', 'MDO') >>> get_scenario_inputs_schema(scenario)
- gemseo.api.get_scenario_options_schema(scenario_type, output_json=False, pretty_print=False)[source]¶
Return the schema of the options of a scenario.
- Parameters:
- Returns:
The schema of the options of the scenario.
- Return type:
Examples
>>> from gemseo.api import get_scenario_options_schema >>> get_scenario_options_schema('MDO')
- gemseo.api.get_surrogate_options_schema(surrogate_name, output_json=False, pretty_print=False)[source]¶
Return the available options for a surrogate discipline.
- Parameters:
- Returns:
The schema of the options of the surrogate discipline.
- Return type:
Examples
>>> from gemseo.api import get_surrogate_options_schema >>> tmp = get_surrogate_options_schema('LinRegSurrogateDiscipline', >>> pretty_print=True)
See also
- gemseo.api.import_discipline(file_path, cls=None)[source]¶
Import a discipline from a pickle file.
- Parameters:
file_path (str | Path) – The path to the file containing the discipline saved with the method
MDODiscipline.serialize().cls (type[MDODiscipline] | None) – A class of discipline. If
None, useMDODiscipline.
- Returns:
The discipline.
- Return type:
- gemseo.api.load_dataset(dataset, **options)[source]¶
Instantiate a dataset.
Typically, benchmark datasets can be found in
gemseo.core.dataset.- Parameters:
dataset (str) – The name of the dataset (its class name).
**options (Any) – The options for creating the dataset.
- Returns:
The dataset.
- Return type:
See also
- gemseo.api.monitor_scenario(scenario, observer)[source]¶
Add an observer to a scenario.
The observer must have an
updatemethod that handles the execution status change of an atom. update(atom) is called everytime an atom execution changes.- Parameters:
scenario (Scenario) – The scenario to monitor.
observer – The observer that handles an update of status.
- Return type:
None
- gemseo.api.print_configuration()[source]¶
Print the current configuration.
The log message contains the successfully loaded modules and failed imports with the reason.
Examples
>>> from gemseo.api import print_configuration >>> print_configuration()
- Return type:
None
- gemseo.api.read_design_space(file_path, header=None)[source]¶
Read a design space from a file.
The following columns must be in the file: “name”, “lower_bound” and “upper_bound”.
- Parameters:
file_path (str | Path) – The path to the text file; the file shall contain space-separated values (the number of spaces is not important) with a row for each variable and at least the bounds of the variable.
header (str | None) – The names of the fields saved in the file. If
None, read them in fhe first row of the file.
- Returns:
The design space.
- Return type:
Examples
>>> from gemseo.api import (create_design_space, export_design_space, >>> read_design_space) >>> source_design_space = create_design_space() >>> source_design_space.add_variable('x', l_b=-1, value=0., u_b=1.) >>> export_design_space(source_design_space, 'file.txt') >>> read_design_space = read_design_space('file.txt') >>> print(read_design_space) Design Space: +------+-------------+-------+-------------+-------+ | name | lower_bound | value | upper_bound | type | +------+-------------+-------+-------------+-------+ | x | -1 | 0 | 1 | float | +------+-------------+-------+-------------+-------+
See also
Examples using create_dataset¶
Examples using create_design_space¶
Pareto front on Binh and Korn problem using a BiLevel formulation
Application: Sobieski’s Super-Sonic Business Jet (MDO)
Example for exterior penalty applied to the Sobieski test case.
Examples using create_discipline¶

Pareto front on Binh and Korn problem using a BiLevel formulation

Application: Sobieski’s Super-Sonic Business Jet (MDO)

MDO formulations for a toy example in aerostructure
Simple disciplinary DOE example on the Sobieski SSBJ test case

Example for exterior penalty applied to the Sobieski test case.
Examples using create_mda¶
Examples using create_parameter_space¶
Examples using create_scalable¶
Examples using create_scenario¶

Pareto front on Binh and Korn problem using a BiLevel formulation
Solve a 2D short cantilever topology optimization problem

Application: Sobieski’s Super-Sonic Business Jet (MDO)

MDO formulations for a toy example in aerostructure

Simple disciplinary DOE example on the Sobieski SSBJ test case

Example for exterior penalty applied to the Sobieski test case.
Examples using create_surrogate¶
Examples using execute_algo¶
Examples using execute_post¶
Examples using export_design_space¶
Examples using generate_coupling_graph¶
Examples using generate_n2_plot¶

MDO formulations for a toy example in aerostructure
Examples using get_algorithm_options_schema¶
Examples using get_available_disciplines¶
Examples using get_available_doe_algorithms¶
Examples using get_available_formulations¶

Application: Sobieski’s Super-Sonic Business Jet (MDO)

Example for exterior penalty applied to the Sobieski test case.